6HR2
 
 | | Crystal structure of PROTAC 2 in complex with the bromodomain of human SMARCA4 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
6HAY
 
 | | Crystal structure of PROTAC 1 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[2-[2-[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]ethoxy]ethoxy]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
6HAX
 
 | | Crystal structure of PROTAC 2 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
2R2S
 
 | | Co(III)bleomycinB2 bound to d(ATTAGTTATAACTAAT) complexed with MMLV RT catalytic fragment | | Descriptor: | BLEOMYCIN B2, COBALT (III) ION, DNA (5'-D(*DAP*DTP*DTP*DAP*DGP*DTP*DT)-3'), ... | | Authors: | Goodwin, K.D, Lewis, M.A, Long, E.C, Georgiadis, M.M. | | Deposit date: | 2007-08-27 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of DNA-bound Co(III) bleomycin B2: Insights on intercalation and minor groove binding.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1RYQ
 
 | | Putative DNA-directed RNA polymerase, subunit e'' from Pyrococcus Furiosus Pfu-263306-001 | | Descriptor: | DNA-directed RNA polymerase, subunit e'', ZINC ION | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Arendall III, W.B, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Parameter-space screening: a powerful tool for high-throughput crystal structure determination.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2R8I
 
 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DC)-3'), ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
2R8H
 
 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DC)-3'), ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
2R8G
 
 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DT)-3', 5'-D(*DTP*DCP*DAP*DCP*(P)P*DGP*DAP*DAP*DAP*DTP*DCP*DCP*DTP*DTP*DCP*DCP*DCP*DCP*DC)-3', ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
8Q3R
 
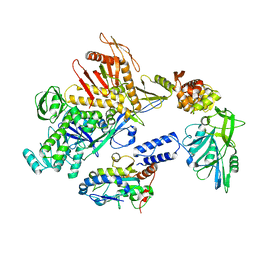 | | Cryo-EM structure of the DNA polymerase holoenzyme E9-A20-D4 of vaccinia virus | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component OPG148, Uracil-DNA glycosylase | | Authors: | Burmeister, W.P, Ballandras-Colas, A, Boettcher, B, Grimm, C. | | Deposit date: | 2023-08-04 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and flexibility of the DNA polymerase holoenzyme of vaccinia virus.
Plos Pathog., 20, 2024
|
|
2HGH
 
 | |
1KP7
 
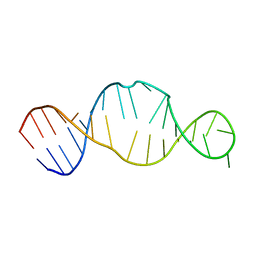 | | Conserved RNA Structure within the HCV IRES eIF3 Binding Site | | Descriptor: | Hepatitis C Virus Internal Ribosome Entry Site Fragment | | Authors: | Gallego, J, Klinck, R, Collier, A.J, Cole, P.T, Harris, S.J, Harrison, G.P, Aboul-ela, F, Walker, S, Varani, G. | | Deposit date: | 2001-12-29 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conserved RNA structure within the HCV IRES eIF3-binding site.
Nat.Struct.Biol., 9, 2002
|
|
2KDQ
 
 | |
5M9D
 
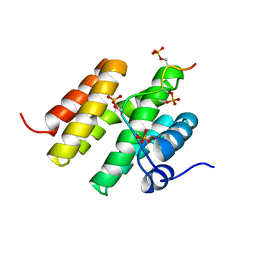 | |
7OMA
 
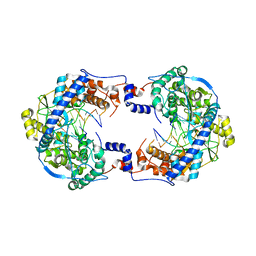 | | Thosea asigna virus RdRP domain elongation complex | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, RNA (5'-R(P*AP*AP*AP*UP*UP*UP*U)-3'), ... | | Authors: | Ferrero, D.S, Falqui, M, Verdaguer, N. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of a Non-Canonical RdRP in Action.
Viruses, 13, 2021
|
|
7OM2
 
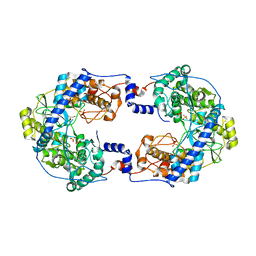 | | Thosea asigna virus RdRP domain in complex with Mg+2 | | Descriptor: | GLYCEROL, MAGNESIUM ION, RNA-dependent RNA polymerase, ... | | Authors: | Ferrero, D.S, Falqui, M, Verdaguer, N. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Snapshots of a Non-Canonical RdRP in Action.
Viruses, 13, 2021
|
|
7Q2J
 
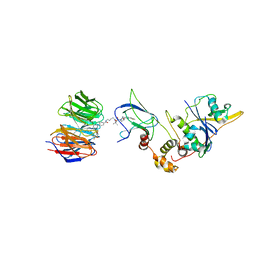 | | Quaternary Complex of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC Homer | | Descriptor: | Elongin-B, Elongin-C, N-[5-[4-[[5-[[(2S)-3,3-dimethyl-1-[(2S,4R)-2-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methylcarbamoyl]-4-oxidanyl-pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]amino]-5-oxidanylidene-pentyl]carbamoyl]phenyl]-2-(4-methylpiperazin-1-yl)phenyl]-6-oxidanylidene-4-(trifluoromethyl)-1H-pyridine-3-carboxamide, ... | | Authors: | Kraemer, A, Doelle, A, Schwalm, M.P, Adhikari, B, Wolf, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-10-25 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
6FI7
 
 | | E.coli Sigma factor S (RpoS) Region 4 | | Descriptor: | RNA polymerase sigma factor RpoS | | Authors: | Liu, B, Matthews, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | T7 phage factor required for managing RpoS inEscherichia coli.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2L41
 
 | | Nab3 RRM - UCUU complex | | Descriptor: | RNA (5'-R(P*UP*CP*UP*U)-3'), RRM domain from Nuclear polyadenylated RNA-binding protein 3 | | Authors: | Stefl, R, Pergoli, R, Hobor, F, Kubicek, K, Zimmermann, M, Pasulka, J, Hofr, C. | | Deposit date: | 2010-09-28 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein
J.Biol.Chem., 286, 2011
|
|
2MQT
 
 | |
2MAO
 
 | |
7Q72
 
 | | Structure of Pla1 in complex with Red1 | | Descriptor: | NURS complex subunit red1, Poly(A) polymerase pla1 | | Authors: | Soni, K, Wild, K, Sinning, I. | | Deposit date: | 2021-11-09 | | Release date: | 2022-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
7Q73
 
 | | Structure of Pla1 apo | | Descriptor: | GLYCEROL, Poly(A) polymerase pla1 | | Authors: | Soni, K, Wild, K, Sinning, I. | | Deposit date: | 2021-11-09 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
7Q74
 
 | |
4OR5
 
 | | Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4 | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Gu, J, Babayeva, N.D, Suwa, Y, Baranovskiy, A.G, Price, D.H, Tahirov, T.H. | | Deposit date: | 2014-02-10 | | Release date: | 2014-04-16 | | Last modified: | 2014-06-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4.
Cell Cycle, 13, 2014
|
|
6YWO
 
 | | CutA in complex with A3 RNA | | Descriptor: | CHLORIDE ION, CutA, MAGNESIUM ION, ... | | Authors: | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
