3MRS
 
 | |
3MRT
 
 | |
3MRU
 
 | | Crystal Structure of Aminoacylhistidine Dipeptidase from Vibrio alginolyticus | | Descriptor: | Aminoacyl-histidine dipeptidase, ZINC ION | | Authors: | Chang, C.-Y, Hsieh, Y.-C, Wu, T.-K, Chen, C.-J. | | Deposit date: | 2010-04-29 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and mutational analysis of aminoacylhistidine dipeptidase from vibrio alginolyticus reveal a new architecture of M20 metallopeptidases
J.Biol.Chem., 285, 2010
|
|
3MRV
 
 | |
3MRW
 
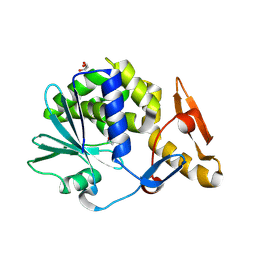 | | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution
To be Published
|
|
3MRX
 
 | |
3MRY
 
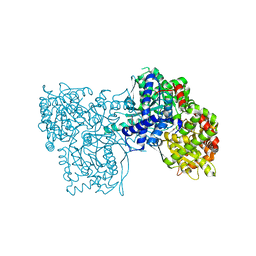
 | | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina with 6-aminopurine at 2.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina with 6-aminopurine at 2.0A resolution
To be Published
|
|
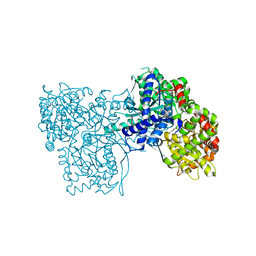
3MS2
 
 | |
3MS3
 
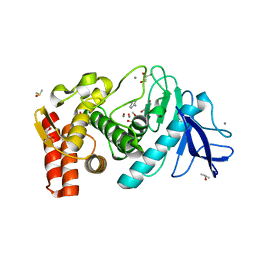 | | Crystal structure of Thermolysin in complex with Aniline | | Descriptor: | ANILINE, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3MS4
 
 | |
3MS5
 
 | | Crystal Structure of Human gamma-butyrobetaine,2-oxoglutarate dioxygenase 1 (BBOX1) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-carboxyethyl)-1,1,1-trimethyldiazanium, Gamma-butyrobetaine dioxygenase, ... | | Authors: | Krojer, T, Kochan, G, McDonough, M.A, von Delft, F, Leung, I.K.H, Henry, L, Claridge, T.D.W, Pilka, E, Ugochukwu, E, Muniz, J, Filippakopoulos, P, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and mechanistic studies on gamma-butyrobetaine hydroxylase.
Chem. Biol., 17, 2010
|
|
3MS6
 
 | | Crystal structure of Hepatitis B X-Interacting Protein (HBXIP) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Hepatitis B virus X-interacting protein, ... | | Authors: | Garcia-Saez, I, Skoufias, D. | | Deposit date: | 2010-04-29 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural Characterization of HBXIP: The Protein That Interacts with the Anti-Apoptotic Protein Survivin and the Oncogenic Viral Protein HBx.
J.Mol.Biol., 405, 2011
|
|
3MS7
 
 | |
3MS8
 
 | | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. | | Descriptor: | GLYCEROL, Intra-cellular xylanase ixt6, SODIUM ION | | Authors: | Solomon, V, Zolotnitsky, G, Alhadeff, R, Shoham, Y, Shoham, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus.
TO BE PUBLISHED
|
|
3MS9
 
 | | ABL kinase in complex with imatinib and a fragment (FRAG1) in the myristate pocket | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, Tyrosine-protein kinase ABL1, ... | | Authors: | Cowan-Jacob, S.W, Rummel, G, Fendrich, G. | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding or bending: distinction of allosteric Abl kinase agonists from antagonists by an NMR-based conformational assay.
J.Am.Chem.Soc., 132, 2010
|
|
3MSA
 
 | | Crystal structure of Thermolysin in complex with 3-Bromophenol | | Descriptor: | 3-bromophenol, CALCIUM ION, GLYCEROL, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3MSC
 
 | |
3MSD
 
 | | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. | | Descriptor: | ACETATE ION, GLYCEROL, Intra-cellular xylanase ixt6, ... | | Authors: | Solomon, V, Zolotnitsky, G, Alhadeff, R, Shoham, Y, Shoham, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus.
TO BE PUBLISHED
|
|
3MSE
 
 | | Crystal structure of C-terminal domain of PF110239. | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase, putative, ... | | Authors: | Wernimont, A.K, Artz, J.D, Hutchinson, A, Sullivan, H, Weadge, J, Tempel, W, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Lin, Y.H, Neculai, A.M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of C-terminal domain of PF110239
To be Published
|
|
3MSF
 
 | | Crystal structure of Thermolysin in complex with Urea | | Descriptor: | CALCIUM ION, GLYCEROL, Thermolysin, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3MSG
 
 | | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. | | Descriptor: | ACETATE ION, GLYCEROL, Intra-cellular xylanase ixt6, ... | | Authors: | Solomon, V, Zolotnitsky, G, Alhadeff, R, Shoham, Y, Shoham, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus.
TO BE PUBLISHED
|
|
3MSH
 
 | |
3MSI
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 | | Descriptor: | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 | | Authors: | Deluca, C.I, Davies, P.L, Ye, Q, Jia, Z. | | Deposit date: | 1997-09-17 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The effects of steric mutations on the structure of type III antifreeze protein and its interaction with ice.
J.Mol.Biol., 275, 1998
|
|
3MSJ
 
 | | Structure of bace (beta secretase) in complex with inhibitor | | Descriptor: | 3-(2-amino-5-chloro-1H-benzimidazol-1-yl)propan-1-ol, BETA-SECRETASE 1, GLYCEROL | | Authors: | Madden, J, Kramer, J, Smith, M.A, Barker, J, Godemann, R. | | Deposit date: | 2010-04-29 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3MSK
 
 | | Fragment Based Discovery and Optimisation of BACE-1 Inhibitors | | Descriptor: | 4-(2-amino-5-chloro-1H-benzimidazol-1-yl)-N-cyclohexyl-N-methylbutanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Smith, M.A, Madden, J.M, Barker, J, Godemann, R, Kraemer, J, Hallett, D. | | Deposit date: | 2010-04-29 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
