4Q0L
 
 | | Crystal structure of catalytic domain of human carbonic anhydrase isozyme XII with inhibitor | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-02 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4PZH
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2,3,5,6-tetrafluoro-4[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
6EPT
 
 | | The ATAD2 bromodomain in complex with compound 12 | | Descriptor: | (2~{R})-~{N}-[5-(5-azanylpyridin-3-yl)-4-ethanoyl-1,3-thiazol-2-yl]piperazine-2-carboxamide, (2~{S})-~{N}-[5-(5-azanylpyridin-3-yl)-4-ethanoyl-1,3-thiazol-2-yl]piperazine-2-carboxamide, ATPase family AAA domain-containing protein 2, ... | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
4PYX
 
 | | Crystal structure of human carbonic anhydrase isozyme II with inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-(cyclooctylamino)-3,5,6-trifluoro-4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-03-28 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
6EPJ
 
 | | The ATAD2 bromodomain in complex with compound 6 | | Descriptor: | (2~{R})-2-azanyl-~{N}-[4-ethanoyl-5-(3-hydroxyphenyl)-1,3-thiazol-2-yl]propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-11 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
4Q09
 
 | | Crystal structure of chimeric carbonic anhydrase XII with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4Q07
 
 | | Crystal structure of chimeric carbonic anhydrase IX with inhibitor | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4Q06
 
 | | Crystal structure of chimeric carbonic anhydrase IX with inhibitor | | Descriptor: | 2-(cyclooctylamino)-3,5,6-trifluoro-4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
6EPV
 
 | | The ATAD2 bromodomain in complex with compound 5 | | Descriptor: | (2~{R})-2-azanyl-~{N}-(4-oxidanylidene-6,7-dihydro-5~{H}-1,3-benzothiazol-2-yl)propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
6EPX
 
 | | The ATAD2 bromodomain in complex with compound 3 | | Descriptor: | (2~{R})-2-carbamimidamido-~{N}-(4-ethanoyl-1,3-thiazolidin-2-yl)propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
6EPU
 
 | | The ATAD2 bromodomain in complex with compound 2 | | Descriptor: | (2~{S})-~{N}-(4-ethanoyl-1,3-thiazol-2-yl)piperazine-2-carboxamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
4PCI
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with B16 | | Descriptor: | (4S)-1-methyl-4-phenyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-04-15 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of BRD4 bromodomain inhibitors by fragment-based high-throughput docking.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6FGI
 
 | | Crystal Structure of BAZ2A bromodomain in complex with 3-amino-2-methylpyridine derivative 2 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[(3~{S})-1,1-bis(oxidanylidene)thian-3-yl]-2-methyl-pyridin-3-amine | | Authors: | Dalle Vedove, A, Marchand, J.-R, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FGH
 
 | | Crystal Structure of BAZ2A bromodomain in complex with 3-amino-2-methylpyridine derivative 1 | | Descriptor: | 2-methyl-~{N}-[(2~{R})-1-methylsulfonylpropan-2-yl]pyridin-3-amine, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Marchand, J.-R, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FGL
 
 | | Crystal Structure of BAZ2A bromodomain in complex with acetylindole compound UZH47 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, MAGNESIUM ION, N-(1-acetyl-1H-indol-3-yl)-N-(5-hydroxy-2-methylphenyl)acetamide | | Authors: | Dalle Vedove, A, Unzue, A, Nevado, C, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
4PYY
 
 | | Crystal structure of human carbonic anhydrase isozyme II with inhibitor | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-03-28 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4TWO
 
 | | Human EphA3 Kinase domain in complex with compound 164 | | Descriptor: | 5-{[3-carbamoyl-4-(3,4-dimethylphenyl)-5-methylthiophen-2-yl]amino}-5-oxopentanoic acid, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-07-01 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural Analysis of the Binding of Type I, I1/2, and II Inhibitors to Eph Tyrosine Kinases.
Acs Med.Chem.Lett., 6, 2015
|
|
8GU0
 
 | | Crystal structure of a fungal halogenase RadH | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Non-heme halogenase radH, ... | | Authors: | Jiang, S.M, Brown, C.J. | | Deposit date: | 2022-09-09 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Further Characterization of Fungal Halogenase RadH and Its Homologs.
Biomolecules, 13, 2023
|
|
4M98
 
 | |
1QUN
 
 | | X-RAY STRUCTURE OF THE FIMC-FIMH CHAPERONE ADHESIN COMPLEX FROM UROPATHOGENIC E.COLI | | Descriptor: | MANNOSE-SPECIFIC ADHESIN FIMH, PAPD-LIKE CHAPERONE FIMC | | Authors: | Choudhury, D, Thompson, A, Stojanoff, V, Langerman, S, Pinkner, J, Hultgren, S.J, Knight, S. | | Deposit date: | 1999-07-01 | | Release date: | 1999-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli.
Science, 285, 1999
|
|
7SZO
 
 | | Structure of a bacterial fimbrial tip containing FocH | | Descriptor: | Chaperone protein FimC, FimF protein, FimG, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Aprikian, P, Sokurenko, E.V. | | Deposit date: | 2021-11-29 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recombinant FimH Adhesin Demonstrates How the Allosteric Catch Bond Mechanism Can Support Fast and Strong Bacterial Attachment in the Absence of Shear.
J.Mol.Biol., 434, 2022
|
|
1KLF
 
 | | FIMH ADHESIN-FIMC CHAPERONE COMPLEX WITH D-MANNOSE | | Descriptor: | CHAPERONE PROTEIN FIMC, FIMH PROTEIN, alpha-D-mannopyranose | | Authors: | Hung, C.S, Bouckaert, J. | | Deposit date: | 2001-12-11 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection.
Mol.Microbiol., 44, 2002
|
|
1KIU
 
 | |
5GHU
 
 | | Crystal structure of YadV chaperone at 1.63 Angstrom | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Pandey, N.K, Bhavesh, N.S. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the usher chaperone YadV reveals a monomer with the proline lock in closed conformation suggestive of an intermediate state.
Febs Lett., 2020
|
|
3RFZ
 
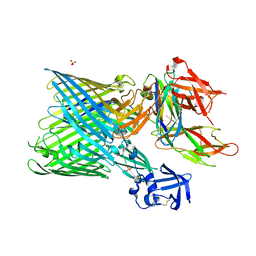 | | Crystal structure of the FimD usher bound to its cognate FimC:FimH substrate | | Descriptor: | Chaperone protein fimC, Outer membrane usher protein, type 1 fimbrial synthesis, ... | | Authors: | Phan, G, Remaut, H, Lebedev, A, Geibel, S, Waksman, G. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
