4HT3
 
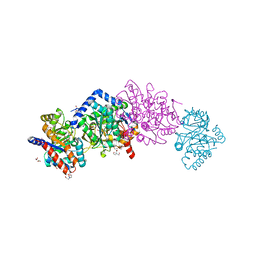 | | The crystal structure of Salmonella typhimurium Tryptophan Synthase at 1.30A complexed with N-(4'-TRIFLUOROMETHOXYBENZENESULFONYL)-2-AMINO-1-ETHYLPHOSPHATE (F9) inhibitor in the alpha site, internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2012-10-31 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
4EB2
 
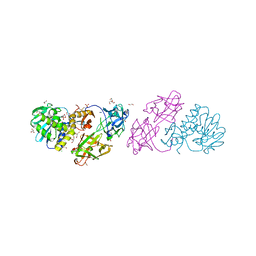 | | Crystal structure Mistletoe Lectin I from Viscum album in complex with n-acetyl-d-glucosamine at 1.94 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Laskov, A.A, Prokofev, I.I, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure Mistletoe Lectin I from Viscum album in complex with N-acetyl-D-glucosamine at 1.94 A resolution.
To be Published
|
|
4I10
 
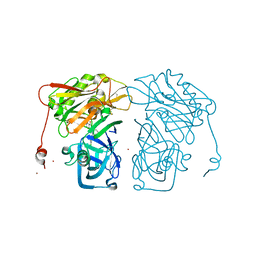 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates | | Descriptor: | 2-{(1S)-1-[(6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)amino]-2-phenylethyl}pyrido[4,3-d]pyrimidin-4(1H)-one, Beta-secretase 1, ZINC ION | | Authors: | Lougheed, J.C, Brecht, E, Yao, N.H. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2YI0
 
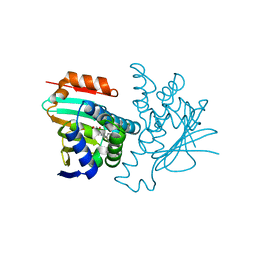 | | Structural characterization of 5-Aryl-4-(5-substituted-2-4- dihydroxyphenyl)-1,2,3-thiadiazole Hsp90 inhibitors. | | Descriptor: | 4-CHLORO-6-[5-(4-METHOXYPHENYL)-1,2,3-THIADIAZOL-4-YL]BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA, MAGNESIUM ION | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2011-05-10 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-Crystalization and in Vitro Biological Characterization of 5-Aryl-4-(5-Substituted-2-4-Dihydroxyphenyl)-1,2,3-Thiadiazole Hsp90 Inhibitors.
Plos One, 7, 2012
|
|
1JLE
 
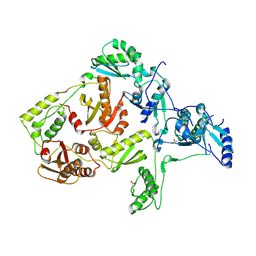 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | HIV-1 RT, A-CHAIN, B-CHAIN | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
3O5S
 
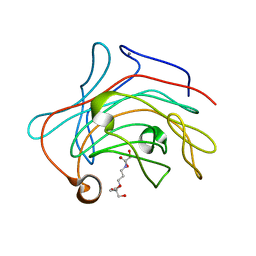 | | Crystal Structure of the endo-beta-1,3-1,4 glucanase from Bacillus subtilis (strain 168) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION | | Authors: | Santos, C.R, Tonoli, C.C.C, Souza, A.R, Furtado, G.P, Ribeiro, L.F, Ward, R.J, Murakami, M.T. | | Deposit date: | 2010-07-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural characterization of a Beta-1,3 1,4-glucanase from Bacillus subtilis 168
PROCESS BIOCHEM, 46, 2011
|
|
1WJE
 
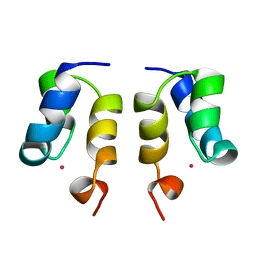 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
4D4B
 
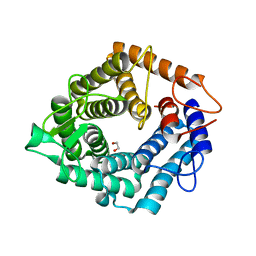 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with MSMSMe | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE, alpha-D-mannopyranose-(1-6)-methyl 1,6-dithio-alpha-D-mannopyranoside | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2CJU
 
 | |
2CHL
 
 | | Structure of casein kinase 1 gamma 3 | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, CASEIN KINASE I ISOFORM GAMMA-3, SULFATE ION | | Authors: | Bunkoczi, G, Salah, E, Rellos, P, Das, S, Fedorov, O, Savitsky, P, Gileadi, O, Sundstrom, M, Edwards, A, Arrowsmith, C, Ugochukwu, E, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-03-15 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Casein Kinase 1 Gamma 3
To be Published
|
|
3SFI
 
 | | Ethionamide Boosters Part 2: Combining Bioisosteric Replacement and Structure-Based Drug Design to Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors. | | Descriptor: | 5,5,5-trifluoro-1-{4-[3-(1,3-thiazol-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}pentan-1-one, Transcriptional regulatory repressor protein (TETR-family) | | Authors: | Flipo, M, Desroses, M, Lecat-Guillet, N, Villemagne, B, Blondiaux, N, Leroux, F, Piveteau, C, Mathys, V, Flament, M.P, Siepmann, J, Villeret, V, Wohlkonig, A, Wintjens, R, Soror, S.H, Christophe, T, Jeon, H.K, Locht, C, Brodin, P, D prez, B, Baulard, A.R, Willand, N. | | Deposit date: | 2011-06-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Ethionamide Boosters. 2. Combining Bioisosteric Replacement and Structure-Based Drug Design To Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors.
J.Med.Chem., 55, 2012
|
|
3OQA
 
 | | Crystal Structures of Multidrug-Resistant Clinical Isolate 769 HIV-1 Protease Variants | | Descriptor: | HIV-1 Protease | | Authors: | Yedidi, R.S, Proteasa, G, Martinez-Cajas, J.L, Vickrey, J.F, Martin, P.D, Wawrzak, Z, Kovari, L.C. | | Deposit date: | 2010-09-02 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3SKF
 
 | |
3OU1
 
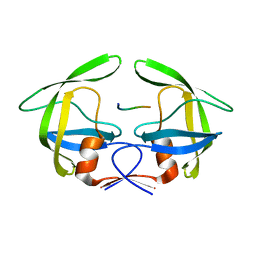 | | MDR769 HIV-1 protease complexed with RH/IN hepta-peptide | | Descriptor: | MDR HIV-1 protease, RH/IN substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
1RDX
 
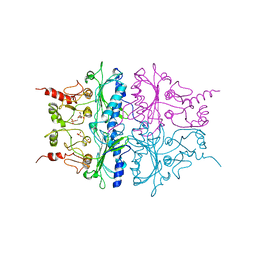 | | R-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
1JLG
 
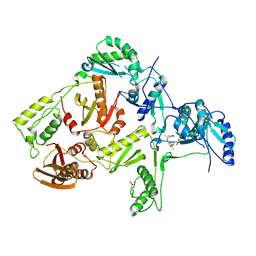 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
2BSZ
 
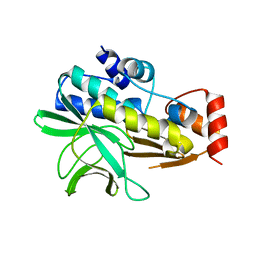 | | Structure of Mesorhizobium loti arylamine N-acetyltransferase 1 | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE 1 | | Authors: | Holton, S.J, Dairou, J, Sandy, J, Rodrigues-Lima, F, Dupret, J.-M, Noble, M.E.M, Sim, E. | | Deposit date: | 2005-05-24 | | Release date: | 2005-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Mesorhizobium Loti Arylamine N-Acetyltransferase 1.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1FP0
 
 | | SOLUTION STRUCTURE OF THE PHD DOMAIN FROM THE KAP-1 COREPRESSOR | | Descriptor: | KAP-1 COREPRESSOR, ZINC ION | | Authors: | Capili, A.D, Schultz, D.C, Rauscher III, F.J, Borden, K.L.B. | | Deposit date: | 2000-08-29 | | Release date: | 2001-01-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains.
EMBO J., 20, 2001
|
|
1FRP
 
 | | CRYSTAL STRUCTURE OF FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE-2,6-BISPHOSPHATE, AMP AND ZN2+ AT 2.0 ANGSTROMS RESOLUTION. ASPECTS OF SYNERGISM BETWEEN INHIBITORS | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Xue, Y, Huang, S, Liang, J.-Y, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-08-26 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 2,6-bisphosphate, AMP, and Zn2+ at 2.0-A resolution: aspects of synergism between inhibitors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
2WIH
 
 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-848125 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, N,1,4,4-TETRAMETHYL-8-{[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]AMINO}-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, ... | | Authors: | Brasca, M.G, Amboldi, N, Ballinari, D, Cameron, A.D, Casale, E, Cervi, G, Colombo, M, Colotta, F, Croci, V, Dalessio, R, Fiorentini, F, Isacchi, A, Mercurio, C, Moretti, W, Panzeri, A, Pastori, W, Pevarello, P, Quartieri, F, Roletto, F, Traquandi, G, Vianello, P, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-05-13 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of N,1,4,4-Tetramethyl-8-{[4-(4-Methylpiperazin-1-Yl)Phenyl]Amino}-4,5-Dihydro-1H-Pyrazolo[4,3-H]Quinazoline-3-Carboxamide (Pha-848125), a Potent, Orally Available Cyclin Dependent Kinase Inhibitor.
J.Med.Chem., 52, 2009
|
|
1FPB
 
 | | CRYSTAL STRUCTURE OF THE NEUTRAL FORM OF FRUCTOSE 1,6-BISPHOSPHATASE COMPLEXED WITH REGULATORY INHIBITOR FRUCTOSE 2,6-BISPHOSPHATE AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Liang, J.Y, Huang, S, Zhang, Y, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the neutral form of fructose 1,6-bisphosphatase complexed with regulatory inhibitor fructose 2,6-bisphosphate at 2.6-A resolution.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1FBP
 
 | | CRYSTAL STRUCTURE OF FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE 6-PHOSPHATE, AMP, AND MAGNESIUM | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Ke, H, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1990-05-31 | | Release date: | 1992-04-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 6-phosphate, AMP, and magnesium.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1GP2
 
 | |
3OU3
 
 | | MDR769 HIV-1 protease complexed with PR/RT hepta-peptide | | Descriptor: | HIV-1 protease, PR/RT substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OUB
 
 | | MDR769 HIV-1 protease complexed with NC/p1 hepta-peptide | | Descriptor: | MDR HIV-1 protease, NC/p1 substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
