3LCB
 
 | | The crystal structure of isocitrate dehydrogenase kinase/phosphatase in complex with its substrate, isocitrate dehydrogenase, from Escherichia coli. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Isocitrate dehydrogenase [NADP], ... | | Authors: | Zheng, J, Jia, Z. | | Deposit date: | 2010-01-10 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bifunctional isocitrate dehydrogenase kinase/phosphatase.
Nature, 465, 2010
|
|
3LDZ
 
 | |
3L9U
 
 | |
3LEI
 
 | | Lectin Domain of Lectinolysin complexed with Fucose | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Platelet aggregation factor Sm-hPAF, ... | | Authors: | Feil, S.C. | | Deposit date: | 2010-01-14 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity.
Structure, 20, 2012
|
|
3LBF
 
 | | Crystal structure of Protein L-isoaspartyl methyltransferase from Escherichia coli | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein-L-isoaspartate O-methyltransferase, ... | | Authors: | Fang, P, Li, X, Wang, J, Niu, L, Teng, M. | | Deposit date: | 2010-01-08 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the protein L-isoaspartyl methyltransferase from Escherichia coli
Cell Biochem.Biophys., 58, 2010
|
|
3LE0
 
 | | Lectin Domain of Lectinolysin complexed with Glycerol | | Descriptor: | CALCIUM ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Feil, S.C. | | Deposit date: | 2010-01-13 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity.
Structure, 20, 2012
|
|
3LGU
 
 | | Y162A mutant of the DegS-deltaPDZ protease | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
3LKQ
 
 | | Crystal Structure of HLA B*3501 in complex with influenza NP418 epitope from 1977 strain | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Kedzierski, L, Valkenburg, S.A, Liu, Y.C, Denholm, J, Richards, M, Rimmelzwaan, G.F, Doherty, P.C, Turner, S.J, Rossjohn, J, Kedzierska, K. | | Deposit date: | 2010-01-27 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3L6X
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin, SULFATE ION | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3LF1
 
 | |
3LGR
 
 | | Xylanase II from Trichoderma reesei cocrystallized with tris-dipicolinate europium | | Descriptor: | EUROPIUM ION, Endo-1,4-beta-xylanase 2, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Pompidor, G, Kahn, R, Maury, O. | | Deposit date: | 2010-01-21 | | Release date: | 2011-01-19 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A dipicolinate lanthanide complex for solving protein structures using anomalous diffraction.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3LF2
 
 | | NADPH Bound Structure of the Short Chain Oxidoreductase Q9HYA2 from Pseudomonas aeruginosa PAO1 Containing an Atypical Catalytic Center | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Huether, R, Umland, T.C, Duax, W.L. | | Deposit date: | 2010-01-15 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The short-chain oxidoreductase Q9HYA2 from Pseudomonas aeruginosa PAO1 contains an atypical catalytic center.
Protein Sci., 19, 2010
|
|
3LJU
 
 | | Crystal structure of full length centaurin alpha-1 bound with the head group of PIP3 | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, Arf-GAP with dual PH domain-containing protein 1, ZINC ION | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-26 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin alpha1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LG6
 
 | |
3LGS
 
 | |
3LPC
 
 | | Crystal structure of a subtilisin-like protease | | Descriptor: | ACETATE ION, AprB2, CALCIUM ION, ... | | Authors: | Porter, C.J, Wong, W, Whisstock, J.C, Rood, J.I, Kennan, R.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
Plos Pathog., 6, 2010
|
|
3L5N
 
 | |
3LIE
 
 | |
3LK5
 
 | | Crystal structure of putative Geranylgeranyl pyrophosphate synthase from Corynebacterium glutamicum | | Descriptor: | GLYCEROL, Geranylgeranyl pyrophosphate synthase | | Authors: | Malashkevich, V.N, Toro, R, Patskovsky, Y, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-27 | | Release date: | 2010-03-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative Geranylgeranyl pyrophosphate synthase from Corynebacterium glutamicum Atcc 13032
To be Published
|
|
3LL1
 
 | |
3L9X
 
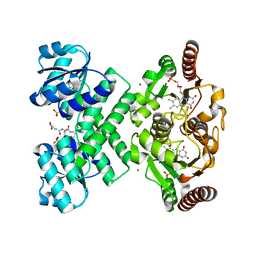 | | KefC C-terminal domain in complex with KefF and ESG | | Descriptor: | FLAVIN MONONUCLEOTIDE, Glutathione-regulated potassium-efflux system protein kefC, linker, ... | | Authors: | Roosild, T.P. | | Deposit date: | 2010-01-05 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of ligand-gated potassium efflux in bacterial pathogens.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LPV
 
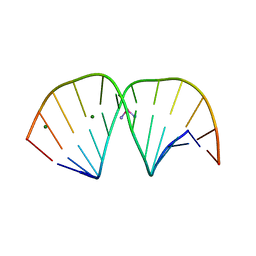 | | X-ray crystal structure of duplex DNA containing a cisplatin 1,2-d(GpG) intrastrand cross-link | | Descriptor: | 5'-D(*CP*CP*TP*CP*TP*GP*GP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3', Cisplatin, ... | | Authors: | Todd, R.C, Lippard, S.J. | | Deposit date: | 2010-02-06 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of duplex DNA containing the cisplatin 1,2-{Pt(NH(3))(2)}(2+)-d(GpG) cross-link at 1.77A resolution.
J.Inorg.Biochem., 104, 2010
|
|
3L5S
 
 | |
3LFF
 
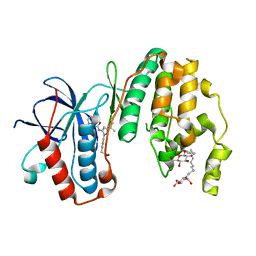 | | Human p38 MAP Kinase in Complex with RL166 | | Descriptor: | (4-{3-tert-butyl-5-[(1,3-thiazol-2-ylcarbamoyl)amino]-1H-pyrazol-1-yl}phenyl)acetic acid, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3LGH
 
 | |
