1HW1
 
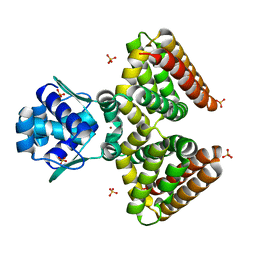 | | THE FADR-DNA COMPLEX: TRANSCRIPTIONAL CONTROL OF FATTY ACID METABOLISM IN ESCHERICHIA COLI | | Descriptor: | FATTY ACID METABOLISM REGULATOR PROTEIN, SULFATE ION, ZINC ION | | Authors: | Xu, Y, Heath, R.J, Li, Z, Rock, C.O, White, S.W. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
2L8N
 
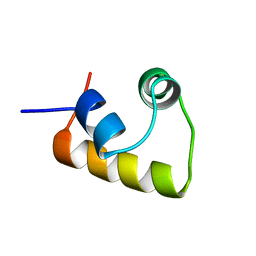 | |
4LDX
 
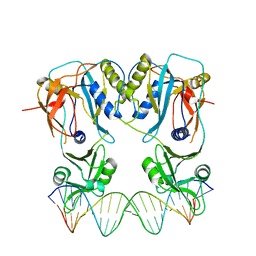 | | Crystal structure of the DNA binding domain of arabidopsis thaliana auxin response factor 1 (ARF1) in complex with protomor-like sequence ER7 | | Descriptor: | Auxin response factor 1, ER7, forward sequence, ... | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
1KB4
 
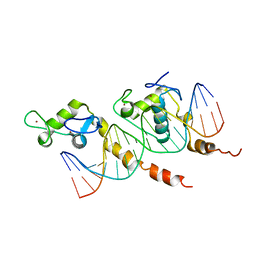 | |
1KB6
 
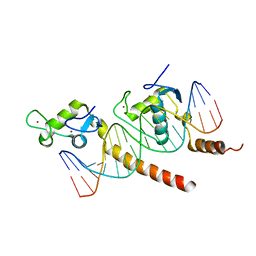 | |
1KB2
 
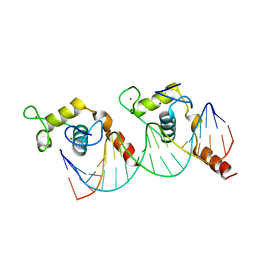 | |
2GDA
 
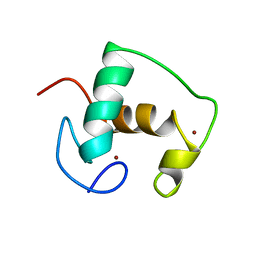 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | Deposit date: | 1994-03-15 | | Release date: | 1994-06-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
2ERE
 
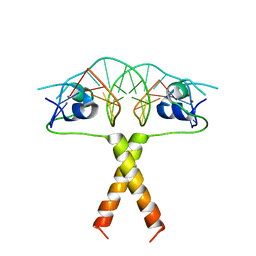 | |
2ERG
 
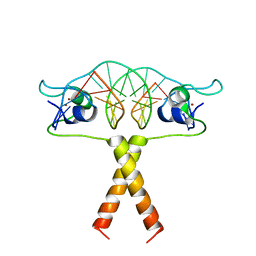 | |
2ER8
 
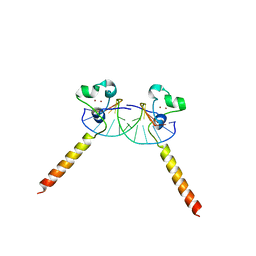 | |
3NFI
 
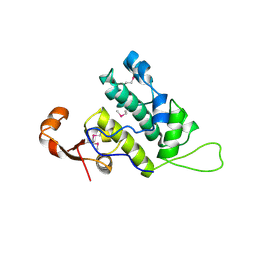 | | Crystal structure of tandem winged helix domain of RNA polymerase I subunit A49 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DNA-directed RNA polymerase I subunit RPA49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
3LAP
 
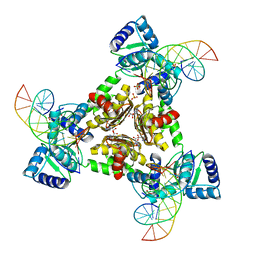 | | The Structure of the Intermediate Complex of the Arginine Repressor from Mycobacterium tuberculosis Bound to its DNA Operator and L-canavanine. | | Descriptor: | 5'-D(*TP*TP*GP*CP*AP*TP*AP*AP*CP*GP*AP*TP*GP*CP*AP*A)-3', 5'-D(*TP*TP*GP*CP*AP*TP*CP*GP*TP*TP*AP*TP*GP*CP*AP*A)-3', Arginine repressor, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-01-06 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | crystal structure of the intermediate complex of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator reveals detailed mechanism of arginine repression.
J.Mol.Biol., 399, 2010
|
|
1LLM
 
 | |
8RAS
 
 | | Plastid-encoded RNA polymerase transcription elongation complex | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
3EXL
 
 | | Crystal Structure of a p53 Core Tetramer Bound to DNA | | Descriptor: | 5'-D(*DGP*DAP*DGP*DCP*DAP*DTP*DGP*DCP*DTP*DCP*DA)-3', 5'-D(*DTP*DTP*DGP*DAP*DGP*DCP*DAP*DTP*DGP*DCP*DTP*DC)-3', CITRATE ANION, ... | | Authors: | Malecka, K.A. | | Deposit date: | 2008-10-16 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a p53 core tetramer bound to DNA.
Oncogene, 28, 2009
|
|
8RDJ
 
 | | Plastid-encoded RNA polymerase transcription elongation complex (Integrated model) | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
1D8G
 
 | | ULTRAHIGH RESOLUTION CRYSTAL STRUCTURE OF B-DNA DECAMER D(CCAGTACTGG) | | Descriptor: | 5'-D(*CP*CP*AP*GP*TP*AP*CP*TP*GP*GP*)-3', CALCIUM ION | | Authors: | Kielkopf, C.L, Ding, S, Kuhn, P, Rees, D.C. | | Deposit date: | 1999-10-23 | | Release date: | 2000-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.74 Å) | | Cite: | Conformational flexibility of B-DNA at 0.74 A resolution: d(CCAGTACTGG)(2).
J.Mol.Biol., 296, 2000
|
|
4NQA
 
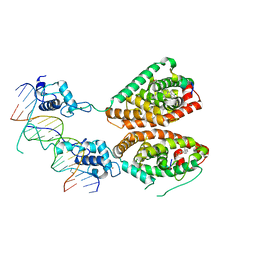 | | Crystal structure of liganded hRXR-alpha/hLXR-beta heterodimer on DNA | | Descriptor: | (9cis)-retinoic acid, 5'-D(*TP*AP*AP*GP*GP*TP*CP*AP*CP*TP*TP*CP*AP*GP*GP*TP*CP*A)-3', 5'-D(*TP*AP*TP*GP*AP*CP*CP*TP*GP*AP*AP*GP*TP*GP*AP*CP*CP*T)-3', ... | | Authors: | Lou, X.H, Toresson, G, Benod, C, Suh, J.H, Phillips, K.J, Webb, P, Gustafsson, J.A. | | Deposit date: | 2013-11-24 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structure of the retinoid X receptor alpha-liver X receptor beta (RXR alpha-LXR beta ) heterodimer on DNA.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3EXJ
 
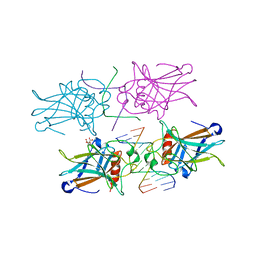 | | Crystal Structure of a p53 Core Tetramer Bound to DNA | | Descriptor: | 5'-D(*DTP*DTP*DGP*DAP*DGP*DCP*DAP*DTP*DGP*DCP*DTP*DC)-3', 5'-D(P*DGP*DAP*DGP*DCP*DAP*DTP*DGP*DCP*DTP*DCP*DA)-3', CITRATE ANION, ... | | Authors: | Malecka, K.A. | | Deposit date: | 2008-10-16 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a p53 core tetramer bound to DNA.
Oncogene, 28, 2009
|
|
2LCV
 
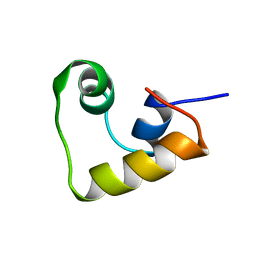 | |
7B0G
 
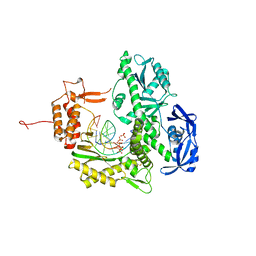 | | TgoT_6G12 binary with 2 hCTPs | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*TP*TP*GP*GP*CP*TP*GP*CP*CP*CP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*GP*CP*AP*GP*()P*())-3'), ... | | Authors: | Samson, C, Legrand, P, Tekpinar, M, Rozenski, J, Abramov, M, Holliger, P, Pinheiro, V, Herdewijn, P, Delarue, M. | | Deposit date: | 2020-11-19 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.
Biomolecules, 10, 2020
|
|
1DPS
 
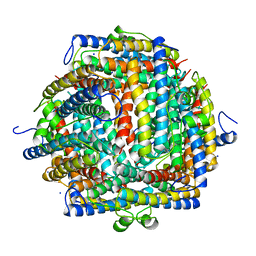 | | THE CRYSTAL STRUCTURE OF DPS, A FERRITIN HOMOLOG THAT BINDS AND PROTECTS DNA | | Descriptor: | DPS, SODIUM ION | | Authors: | Grant, R.A, Filman, D.J, Finkel, S.E, Kolter, R, Hogle, J.M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Dps, a ferritin homolog that binds and protects DNA.
Nat.Struct.Biol., 5, 1998
|
|
7B07
 
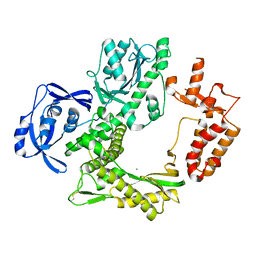 | | TgoT_6G12 apo | | Descriptor: | CALCIUM ION, DNA polymerase | | Authors: | Samson, C, Legrand, P, Tekpinar, M, Rozenski, J, Abramov, M, Holliger, P, Pinheiro, V, Herdewijn, P, Delarue, M. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.
Biomolecules, 10, 2020
|
|
1F9F
 
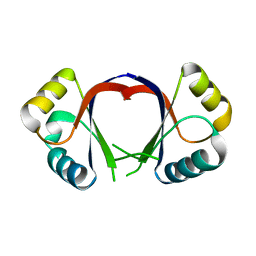 | | CRYSTAL STRUCTURE OF THE HPV-18 E2 DNA-BINDING DOMAIN | | Descriptor: | Regulatory protein E2, SULFATE ION | | Authors: | Kim, S.S, Tam, J, Wang, A.F, Hegde, R. | | Deposit date: | 2000-07-10 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of DNA target discrimination by papillomavirus E2 proteins.
J.Biol.Chem., 275, 2000
|
|
8CWR
 
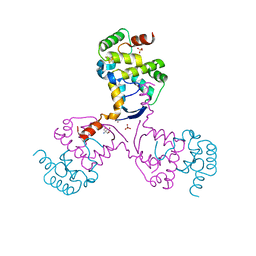 | |
