8YHI
 
 | | The Crystal Structure of SHP1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 6 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Shen, Z. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of SHP1 from Biortus.
To Be Published
|
|
8YHH
 
 | | The Crystal Structure of Mitotic Kinesin Eg5 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Wu, B. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Mitotic Kinesin Eg5 from Biortus
To Be Published
|
|
8YHF
 
 | | The Crystal Structure of the Type I TGF beta receptor from Biortus. | | Descriptor: | 2-fluoranyl-~{N}-[[5-(6-methylpyridin-2-yl)-4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-1~{H}-imidazol-2-yl]methyl]aniline, TGF-beta receptor type-1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Type I TGF beta receptor from Biortus.
To Be Published
|
|
8YGZ
 
 | | The Crystal Structure of TGF beta R2 kinase domain from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, TGF-beta receptor type-2 | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of TGF beta R2 kinase domain from Biortus.
To Be Published
|
|
8YGY
 
 | | Structure of the KLK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Kallikrein-1, SULFATE ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of the KLK1 from Biortus.
To Be Published
|
|
8YGX
 
 | |
8YGW
 
 | | The Crystal Structure of MAPK11 from Biortus | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 11 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Guo, S. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | The Crystal Structure of MAPK11 from Biortus.
To Be Published
|
|
8YGJ
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 28-nt) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 2024
|
|
8YG2
 
 | |
8YFJ
 
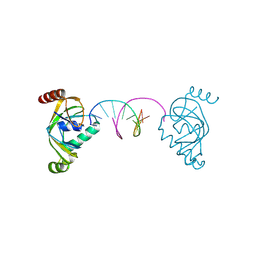 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-ATTGGATCCAAT-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*AP*TP*TP*GP*GP*AP*TP*CP*CP*AP*AP*T)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-24 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 2024
|
|
8YFI
 
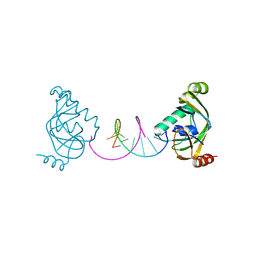 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCA-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-24 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 2024
|
|
8YF1
 
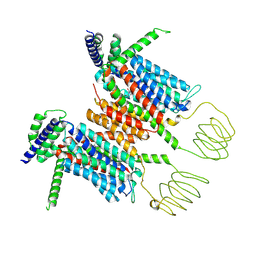 | |
8YF0
 
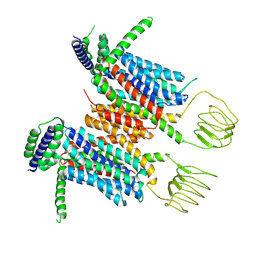 | |
8YEY
 
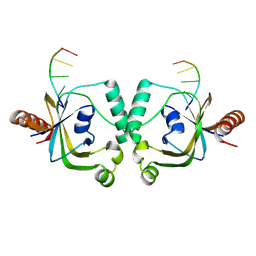 | | TRIP4 ASCH domain in complex with ssDNA-1 | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*GP*TP*TP*TP*C)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-23 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 2024
|
|
8YEW
 
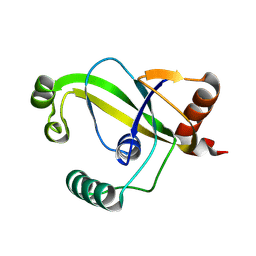 | | TRIP4 ASCH domain in unliganded form | | Descriptor: | Activating signal cointegrator 1 | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-23 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 2024
|
|
8YEA
 
 | |
8YE5
 
 | |
8YE0
 
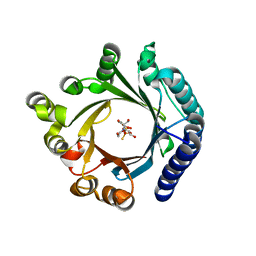 | | Crystal structure of KgpF prenyltransferase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | Authors: | Hamada, K, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2024-02-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | De Novo Discovery of Pseudo-Natural Prenylated Macrocyclic Peptide Ligands.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8YDO
 
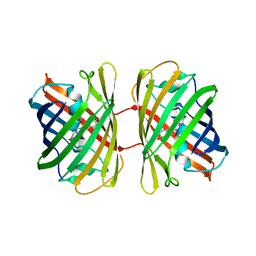 | | Crystal structure of dKeima570 | | Descriptor: | Large stokes shift fluorescent protein | | Authors: | Nam, K.H. | | Deposit date: | 2024-02-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dKeima570
To Be Published
|
|
8YD8
 
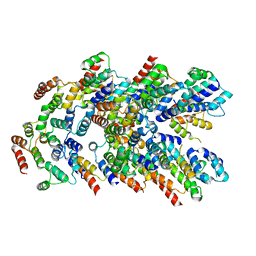 | |
8YD7
 
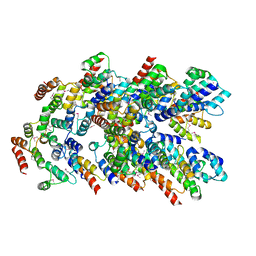 | |
8YCM
 
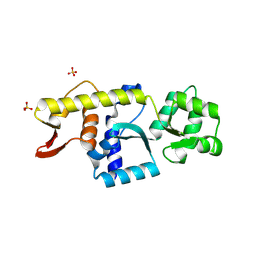 | | Monomeric Human STK19 | | Descriptor: | Isoform 4 of Inactive serine/threonine-protein kinase 19, SULFATE ION | | Authors: | Li, J, Ma, X, Dong, Z. | | Deposit date: | 2024-02-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Mutations found in cancer patients compromise DNA binding of the winged helix protein STK19.
Sci Rep, 14, 2024
|
|
8YC0
 
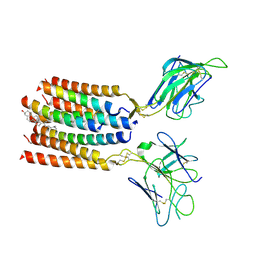 | | T cell receptor V delta2 V gamma9 in GDN | | Descriptor: | CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2024-02-17 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8YBX
 
 | |
8YBH
 
 | |
