1PXW
 
 | | Crystal structure of L7Ae sRNP core protein from Pyrococcus abyssii | | Descriptor: | LSU ribosomal protein L7AE | | Authors: | Charron, C, Manival, X, Charpentier, B, Branlant, C, Aubry, A. | | Deposit date: | 2003-07-07 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Purification, crystallization and preliminary X-ray diffraction data of L7Ae sRNP core protein from Pyrococcus abyssii.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PND
 
 | |
1Q3P
 
 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | Descriptor: | C-terminal hexapeptide from Guanylate kinase-associated protein, Shank1 | | Authors: | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
3QRI
 
 | | The crystal structure of human abl1 kinase domain in complex with DCC-2036 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, SODIUM ION, Tyrosine-protein kinase ABL1 | | Authors: | Chan, W.W, Wise, S.C, Kaufman, M.D, Ahn, Y.M, Ensinger, C.L, Haack, T, Hood, M.M, Jones, J, Lord, J.W, Lu, W.P, Miller, D, Patt, W.C, Smith, B.D, Petillo, P.A, Rutkoski, T.J, Telikepalli, H, Vogeti, L, Yao, T, Chun, L, Clark, R, Evangelista, P, Gavrilescu, L.C, Lazarides, K, Zaleskas, V.M, Stewart, L.J, Van Etten, R.A, Flynn, D.L. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036.
Cancer Cell, 19, 2011
|
|
1ZT7
 
 | | crystal structure of class I MHC H-2Kk in complex with a nonapeptide | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-K alpha chain, ... | | Authors: | Kellenberger, C, Roussel, A, Malissen, B. | | Deposit date: | 2005-05-26 | | Release date: | 2005-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The H-2Kk MHC peptide-binding groove anchors the backbone of an octameric antigenic peptide in an unprecedented mode.
J Immunol., 175, 2005
|
|
3R3D
 
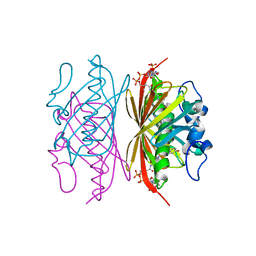 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant T77S complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA thioesterase | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
3R36
 
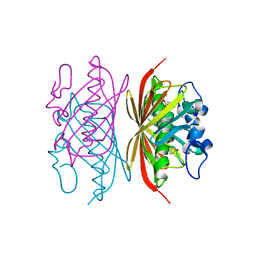 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant E73Q complexed with 4-hydroxybenzoic acid | | Descriptor: | 4-hydroxybenzoyl-CoA thioesterase, P-HYDROXYBENZOIC ACID | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
1XIQ
 
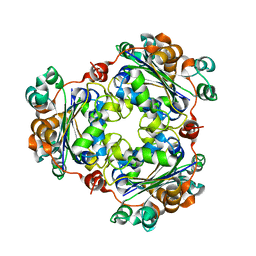 | |
1KF0
 
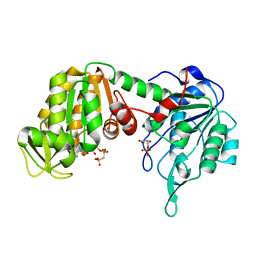 | | Crystal Structure of Pig Muscle Phosphoglycerate Kinase Ternary Complex with AMP-PCP and 3PG | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Kovari, Z, Flachner, B, Naray-Szabo, G, Vas, M. | | Deposit date: | 2001-11-19 | | Release date: | 2002-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and thiol-reactivity studies on the complex of pig muscle phosphoglycerate kinase with ATP analogues: correlation between nucleotide binding mode and helix flexibility.
Biochemistry, 41, 2002
|
|
1NGD
 
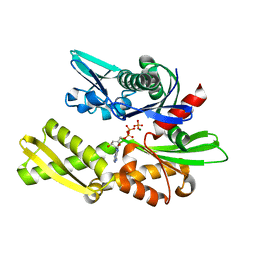 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1KVT
 
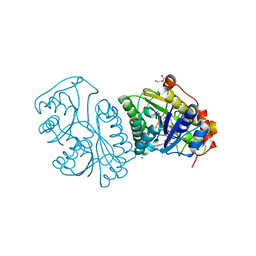 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
1XF6
 
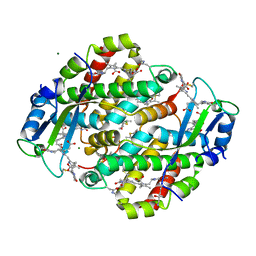 | | High resolution crystal structure of phycoerythrin 545 from the marine cryptophyte rhodomonas CS24 | | Descriptor: | 15,16-DIHYDROBILIVERDIN, B-phycoerythrin beta chain, CHLORIDE ION, ... | | Authors: | Doust, A.B, Marai, C.N.J, Harrop, S.J, Wilk, K.E, Curmi, P.M.G, Scholes, G.D. | | Deposit date: | 2004-09-14 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Developing a structure-function model for the cryptophyte phycoerythrin 545 using ultrahigh resolution crystallography and ultrafast laser spectroscopy
J.Mol.Biol., 344, 2004
|
|
1PLG
 
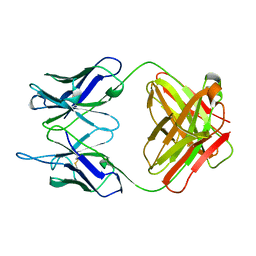 | | EVIDENCE FOR THE EXTENDED HELICAL NATURE OF POLYSACCHARIDE EPITOPES. THE 2.8 ANGSTROMS RESOLUTION STRUCTURE AND THERMODYNAMICS OF LIGAND BINDING OF AN ANTIGEN BINDING FRAGMENT SPECIFIC FOR ALPHA-(2->8)-POLYSIALIC ACID | | Descriptor: | IGG2A=KAPPA= | | Authors: | Evans, S.V, Sigurskjold, B.W, Jennings, H.J, Brisson, J.-R, Tse, W.C, To, R, Altman, E, Frosch, M, Weisgerber, C, Kratzin, H, Klebert, S, Vaesen, M, Bitter-Suermann, D, Rose, D.R, Young, N.M, Bundle, D.R. | | Deposit date: | 1995-04-24 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for the extended helical nature of polysaccharide epitopes. The 2.8 A resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for alpha-(2-->8)-polysialic acid.
Biochemistry, 34, 1995
|
|
1PPA
 
 | | THE CRYSTAL STRUCTURE OF A LYSINE 49 PHOSPHOLIPASE A2 FROM THE VENOM OF THE COTTONMOUTH SNAKE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | ANILINE, PHOSPHOLIPASE A2 | | Authors: | Holland, D.R, Clancy, L.L, Muchmore, S.W, Rydel, T.J, Einspahr, H.M, Finzel, B.C, Heinrikson, R.L, Watenpaugh, K.D. | | Deposit date: | 1991-10-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a lysine 49 phospholipase A2 from the venom of the cottonmouth snake at 2.0-A resolution.
J.Biol.Chem., 265, 1990
|
|
1XG0
 
 | | High resolution crystal structure of phycoerythrin 545 from the marine cryptophyte rhodomonas CS24 | | Descriptor: | 15,16-DIHYDROBILIVERDIN, B-phycoerythrin beta chain, CHLORIDE ION, ... | | Authors: | Doust, A.B, Marai, C.N.J, Harrop, S.J, Wilk, K.E, Curmi, P.M.G, Scholes, G.D. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Developing a structure-function model for the cryptophyte phycoerythrin 545 using ultrahigh resolution crystallography and ultrafast laser spectroscopy
J.Mol.Biol., 344, 2004
|
|
1XU1
 
 | | The crystal structure of APRIL bound to TACI | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
2A93
 
 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, 40 STRUCTURES | | Descriptor: | C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER | | Authors: | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | Deposit date: | 1998-06-09 | | Release date: | 1999-01-27 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
3AT7
 
 | | Crystal structure of bacterial cell-surface alginate-binding protein Algp7 | | Descriptor: | Alginate-binding flagellin | | Authors: | Maruyama, Y, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-12-27 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bacterial cell-surface alginate-binding protein with an M75 peptidase motif.
Biochem.Biophys.Res.Commun., 405, 2011
|
|
2FLQ
 
 | |
1NOL
 
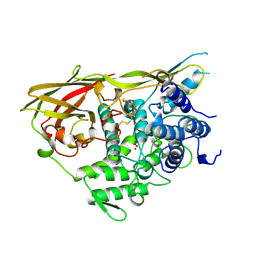 | | OXYGENATED HEMOCYANIN (SUBUNIT TYPE II) | | Descriptor: | CALCIUM ION, COPPER (II) ION, HEMOCYANIN (SUBUNIT TYPE II), ... | | Authors: | Hazes, B, Hol, W.G.J. | | Deposit date: | 1995-10-17 | | Release date: | 1996-03-08 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of deoxygenated Limulus polyphemus subunit II hemocyanin at 2.18 A resolution: clues for a mechanism for allosteric regulation.
Protein Sci., 2, 1993
|
|
1JXT
 
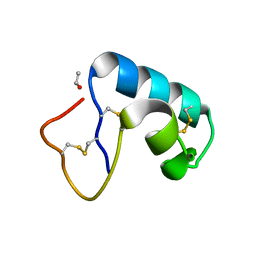 | | CRAMBIN MIXED SEQUENCE FORM AT 160 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-08 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1MSH
 
 | |
1NGC
 
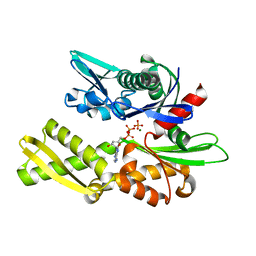 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NGA
 
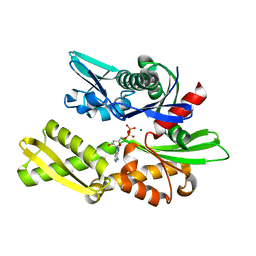 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NGI
 
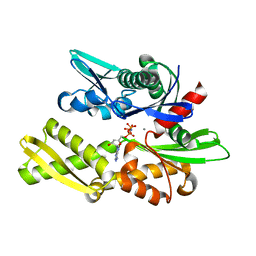 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | CALCIUM ION, HEAT-SHOCK COGNATE 70 kD PROTEIN, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
