1TLA
 
 | | HYDROPHOBIC CORE REPACKING AND AROMATIC-AROMATIC INTERACTION IN THE THERMOSTABLE MUTANT OF T4 LYSOZYME SER 117 (RIGHT ARROW) PHE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, T4 LYSOZYME | | Authors: | Anderson, D.E, Hurley, J.H, Nicholson, H, Baase, W.A, Matthews, B.W. | | Deposit date: | 1993-03-22 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophobic core repacking and aromatic-aromatic interaction in the thermostable mutant of T4 lysozyme Ser 117-->Phe.
Protein Sci., 2, 1993
|
|
1YLJ
 
 | | Atomic resolution structure of CTX-M-9 beta-lactamase | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-lactamase CTX-M-9a | | Authors: | Chen, Y, Delmas, J, Sirot, J, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution Structures of CTX-M beta-Lactamases: Extended Spectrum Activities from Increased Mobility and Decreased Stability.
J.Mol.Biol., 348, 2005
|
|
1YLP
 
 | | Atomic resolution structure of CTX-M-27 beta-lactamase | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-lactamase CTX-M-27 | | Authors: | Chen, Y, Delmas, J, Sirot, J, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic Resolution Structures of CTX-M beta-Lactamases: Extended Spectrum Activities from Increased Mobility and Decreased Stability.
J.Mol.Biol., 348, 2005
|
|
1KVS
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
1KVR
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
1KNT
 
 | | THE 1.6 ANGSTROMS STRUCTURE OF THE KUNITZ-TYPE DOMAIN FROM THE ALPHA3 CHAIN OF THE HUMAN TYPE VI COLLAGEN | | Descriptor: | COLLAGEN TYPE VI, SULFATE ION | | Authors: | Arnoux, B, Merigeau, K, Saludjian, P, Norris, F, Norris, K, Bjorn, S, Olsen, O, Petersen, L, Ducruix, A. | | Deposit date: | 1994-08-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A structure of Kunitz-type domain from the alpha 3 chain of human type VI collagen.
J.Mol.Biol., 246, 1995
|
|
1ZBP
 
 | | X-Ray Crystal Structure of Protein VPA1032 from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR44 | | Descriptor: | hypothetical protein VPA1032 | | Authors: | Forouhar, F, Yong, W, Vorobiev, S.M, Ciao, M, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2EPN
 
 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, N-acetyl-beta-D-glucosaminidase | | Authors: | Langley, D.B. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
2O5B
 
 | | Manganese horse heart myoglobin, reduced | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, Myoglobin, SULFATE ION | | Authors: | Richter-Addo, G.B, Zahran, Z.N, Chooback, L, Copeland, D.M, West, A.H. | | Deposit date: | 2006-12-05 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of manganese- and cobalt-substituted myoglobin in complex with NO and nitrite reveal unusual ligand conformations.
J.Inorg.Biochem., 102, 2008
|
|
2O5L
 
 | | Manganese horse heart myoglobin, methanol modified | | Descriptor: | METHANOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING MN, ... | | Authors: | Richter-Addo, G.B, Zahran, Z.N, Chooback, L, Copeland, D.M, West, A.H. | | Deposit date: | 2006-12-06 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of manganese- and cobalt-substituted myoglobin in complex with NO and nitrite reveal unusual ligand conformations.
J.Inorg.Biochem., 102, 2008
|
|
2O5O
 
 | | Manganese horse heart myoglobin, nitrite modified | | Descriptor: | Myoglobin, NITRITE ION, PROTOPORPHYRIN IX CONTAINING MN, ... | | Authors: | Richter-Addo, G.B, Zahran, Z.N, Chooback, L, Copeland, D.M, West, A.H. | | Deposit date: | 2006-12-06 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of manganese- and cobalt-substituted myoglobin in complex with NO and nitrite reveal unusual ligand conformations.
J.Inorg.Biochem., 102, 2008
|
|
2O5T
 
 | | Cobalt horse heart myoglobin, oxidized | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING CO, SULFATE ION | | Authors: | Richter-Addo, G.B, Zahran, Z.N, Chooback, L, Copeland, D.M, West, A.H. | | Deposit date: | 2006-12-06 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of manganese- and cobalt-substituted myoglobin in complex with NO and nitrite reveal unusual ligand conformations.
J.Inorg.Biochem., 102, 2008
|
|
1I8U
 
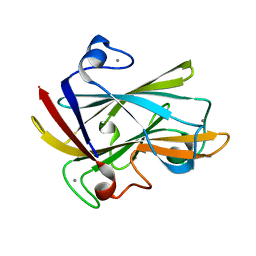 | | FAMILY 9 CARBOHYDRATE-BINDING MODULE FROM THERMOTOGA MARITIMA XYLANASE 10A | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A | | Authors: | Notenboom, V, Boraston, A.B, Warren, R.A.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-03-16 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms.
Biochemistry, 40, 2001
|
|
3R3C
 
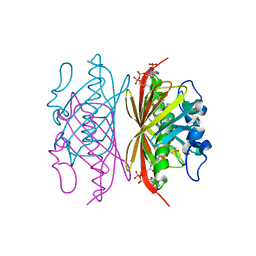 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant H64A complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA thioesterase | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
1TTV
 
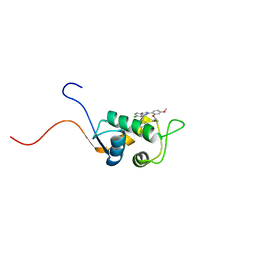 | | NMR Structure of a Complex Between MDM2 and a Small Molecule Inhibitor | | Descriptor: | 1-{[4,5-BIS(4-CHLOROPHENYL)-2-(2-ISOPROPOXY-4-METHOXYPHENYL)-4,5-DIHYDRO-1H-IMIDAZOL-1-YL]CARBONYL}PIPERAZINE, Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Fry, D.C, Emerson, S.D, Palme, S, Vu, B.T, Liu, C.M, Podlaski, F. | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a complex between MDM2 and a small molecule inhibitor.
J.Biomol.Nmr, 30, 2004
|
|
1U96
 
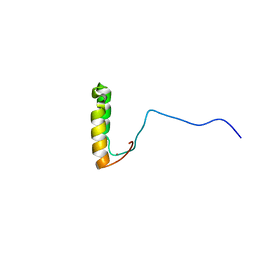 | | Solution Structure of Yeast Cox17 with Copper Bound | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase copper chaperone | | Authors: | Abajian, C, Yatsunyk, L.A, Ramirez, B.E, Rosenzweig, A.C. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Yeast cox17 solution structure and Copper(I) binding.
J.Biol.Chem., 279, 2004
|
|
1JXW
 
 | | CRAMBIN MIXED SEQUENCE FORM AT 180 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3GEN
 
 | | The 1.6 A crystal structure of human bruton's tyrosine kinase bound to a pyrrolopyrimidine-containing compound | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, Tyrosine-protein kinase BTK | | Authors: | Marcotte, D.J, Liu, Y.T, Arduini, R.M, Hession, C.A, Miatkowski, K, Wildes, C.P, Cullen, P.F, Hopkins, B, Mertsching, E, Jenkins, T.J, Romanowski, M.J, Baker, D.P, Silvian, L.F. | | Deposit date: | 2009-02-25 | | Release date: | 2010-01-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggest a mechanism of activation for TEC family kinases.
Protein Sci., 19, 2010
|
|
1JBC
 
 | | CONCANAVALIN A | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Parkin, S, Rupp, B, Hope, H. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structure of concanavalin A at 120 K.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1JBY
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT LOW PH | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
4I98
 
 | |
1RTP
 
 | | REFINED X-RAY STRUCTURE OF RAT PARVALBUMIN, A MAMMALIAN ALPHA-LINEAGE PARVALBUMIN, AT 2.0 A RESOLUTION | | Descriptor: | ALPHA-PARVALBUMIN, CALCIUM ION | | Authors: | Mcphalen, C.A, Sielecki, A.R, Santarsiero, B.D, James, M.N.G. | | Deposit date: | 1993-05-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structure of rat parvalbumin, a mammalian alpha-lineage parvalbumin, at 2.0 A resolution.
J.Mol.Biol., 235, 1994
|
|
2O0I
 
 | | crystal structure of the R185A mutant of the N-terminal domain of the Group B Streptococcus Alpha C protein | | Descriptor: | C protein alpha-antigen | | Authors: | Hogle, J.M, Filman, D.J, Baron, M.J, Madoff, L.C, Iglesias, A. | | Deposit date: | 2006-11-27 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification of a glycosaminoglycan binding region of the alpha C protein that mediates entry of group B streptococci into host cells.
J.Biol.Chem., 282, 2007
|
|
3TS6
 
 | |
1KRJ
 
 | | Engineering Calcium-binding site into Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome c Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bonagura, C.A, Bhaskar, B, Sundaramoorthy, M, Poulos, T.L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conversion of an engineered potassium-binding site into a calcium-selective site in cytochrome c peroxidase.
J.Biol.Chem., 274, 1999
|
|
