5PU2
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 180) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PUM
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 200) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5SFY
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2c(c1nc(nn1c(c2)C)C=Cc3nc(nn3C)c4c[nH]nc4)C, micromolar IC50=0.22176 | | Descriptor: | (4S)-5,8-dimethyl-2-{(E)-2-[1-methyl-3-(1H-pyrazol-4-yl)-1H-1,2,4-triazol-5-yl]ethenyl}[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5PUZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 213) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PVF
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 229) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PVX
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 248) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5SGC
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n3cn1c(nc(n1)N2CCOCC2)cc3NC(c4c(cnn4C)C(N5CCC5)=O)=O, micromolar IC50=0.004831 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[(4S)-2-(morpholin-4-yl)[1,2,4]triazolo[1,5-c]pyrimidin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4QBY
 
 | | yCP in complex with BOC-ALA-ALA-ALA-CHO | | Descriptor: | BOC-ALA-ALA-ALA-CHO, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Arciniega, M, Beck, P, Lange, O, Groll, M, Huber, R. | | Deposit date: | 2014-05-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential global structural changes in the core particle of yeast and mouse proteasome induced by ligand binding.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7H6Q
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1148165337 (CHIKV_MacB-x0312) | | Descriptor: | 2-(1H-indazol-1-yl)-N,N-dimethylacetamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
5SGK
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH [C@@H]2(Cc1nc(cn1CC2)c3ccccc3C)NC(=O)c5c(C(=O)N4CCC4)cnn5C, micromolar IC50=0.007851 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[(4R,7R)-2-(2-methylphenyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Bleicher, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
7QEP
 
 | | Cryo-EM structure of the ribosome from Encephalitozoon cuniculi | | Descriptor: | 18S ribosomal RNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Nicholson, D, Ranson, N.A, Melnikov, S.V. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Adaptation to genome decay in the structure of the smallest eukaryotic ribosome
Nat Commun, 13, 2022
|
|
7H9A
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z56899125 (CHIKV_MacB-x1410) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-fluoro-1,3-benzothiazol-2-amine, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
4N5P
 
 | | Thermolysin in complex with UBTLN20 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2013-10-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Methyl, Ethyl, Propyl, Butyl: Futile But Not for Water, as the Correlation of Structure and Thermodynamic Signature Shows in a Congeneric Series of Thermolysin Inhibitors.
Chemmedchem, 4, 2014
|
|
3HHM
 
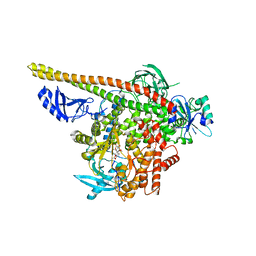 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha and the drug wortmannin | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | Deposit date: | 2009-05-15 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5SE8
 
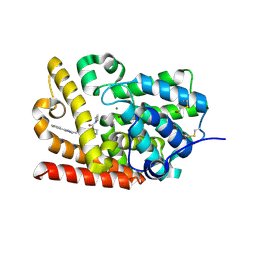 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n1(C)c(nc(n1)c2ccccc2)CCNC(c4c(C(N3CCC3)=O)cnn4C)=O, micromolar IC50=0.021263 | | Descriptor: | 4-(azetidine-1-carbonyl)-2-methyl-N-[2-(1-methyl-3-phenyl-1H-1,2,4-triazol-5-yl)ethyl]-2,5-dihydro-1H-pyrazole-3-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Koerner, M, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
7H6Y
 
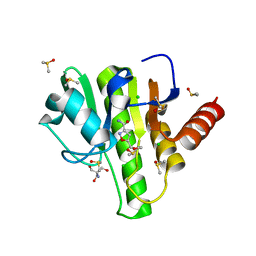 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1270393711 (CHIKV_MacB-x0404) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-(methoxymethyl)-1,2-oxazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
7LRT
 
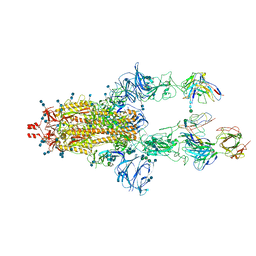 | |
7H71
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1416571195 (CHIKV_MacB-x0441) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[(cyclopent-3-en-1-yl)amino]pyridine-4-carboxamide, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
5SH2
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH C(c1c(cnn1C)C(N2CCC2)=O)(=O)Nc3cc4c(cc3)sc(n4)N5CCOCC5, micromolar IC50=0.001856 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[2-(morpholin-4-yl)-1,3-benzothiazol-5-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4ERT
 
 | |
7H8J
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z362020366 (CHIKV_MacB-x1198) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
5Q0X
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 6-(4-{[3-(3,5-dichloropyridin-4-yl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-2-methylphenyl)-1-methyl-1H-indole-3-carbox ylic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
6JGL
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with methyl 2-thio-beta-sophoroside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
5SFO
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2(cc(c1ccccc1)nc2CCNC(c4c(C(=O)N3CCC3)cnn4C)=O)C, micromolar IC50=0.004013 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethyl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Koerner, M, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
7H8X
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z57169486 (CHIKV_MacB-x1320) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, COUMARIN, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
