5TC3
 
 | | Structure of IMP dehydrogenase from Ashbya gossypii bound to ATP and GDP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Justel, D, de Pereda, J.M, Revuelta, J.L, Buey, R.M. | | Deposit date: | 2016-09-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases.
Sci Rep, 7, 2017
|
|
2Q2Y
 
 | | Crystal Structure of KSP in complex with Inhibitor 1 | | Descriptor: | 1-{(3R,3AR)-3-[3-(4-ACETYLPIPERAZIN-1-YL)PROPYL]-8-FLUORO-3-PHENYL-3A,4-DIHYDRO-3H-PYRAZOLO[5,1-C][1,4]BENZOXAZIN-2-YL}ETHANONE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2007-05-29 | | Release date: | 2007-09-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 8: Design and synthesis of 1,4-diaryl-4,5-dihydropyrazoles as potent inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2SBA
 
 | | SOYBEAN AGGLUTININ COMPLEXED WITH 2,6-PENTASACCHARIDE | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Dessen, A, Gupta, D, Sabesan, S, Brewer, C.F, Sacchettini, J.C. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystal structure of the soybean agglutinin cross-linked with a biantennary analog of the blood group I carbohydrate antigen.
Biochemistry, 34, 1995
|
|
2RFN
 
 | | x-ray structure of c-Met with inhibitor. | | Descriptor: | 2-benzyl-5-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
5UAN
 
 | | Crystal structure of multi-domain RAR-beta-RXR-alpha heterodimer on DNA | | Descriptor: | (9cis)-retinoic acid, DNA (5'-D(*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*AP*G)-3'), ... | | Authors: | Chandra, V, Wu, D, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-12-19 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.508 Å) | | Cite: | The quaternary architecture of RAR beta-RXR alpha heterodimer facilitates domain-domain signal transmission.
Nat Commun, 8, 2017
|
|
4CBA
 
 | | Structural of delta 1-76 CTNNBL1 in space group I222 | | Descriptor: | 1,2-ETHANEDIOL, BETA-CATENIN-LIKE PROTEIN 1, SULFATE ION | | Authors: | Ganesh, K, vanMaldegem, F, Telerman, S.B, Simpson, P, Johnson, C.M, Williams, R.L, Neuberger, M.S, Rada, C. | | Deposit date: | 2013-10-10 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and mutational analysis reveals that CTNNBL1 binds NLSs in a manner distinct from that of its closest armadillo-relative, karyopherin alpha.
Febs Lett., 588, 2014
|
|
3S72
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1H-benzimidazole-2-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3S78
 
 | |
3S77
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1,3-thiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3F6P
 
 | | Crystal Structure of unphosphorelated receiver domain of YycF | | Descriptor: | Transcriptional regulatory protein yycF | | Authors: | Zhao, H, Tang, L. | | Deposit date: | 2008-11-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Preliminary crystallographic studies of the regulatory domain of response regulator YycF from an essential two-component signal transduction system.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
4IJG
 
 | | Crystal structure of monomeric bacteriophytochrome | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Auldridge, M.E. | | Deposit date: | 2012-12-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Origins of fluorescence in evolved bacteriophytochromes.
J.Biol.Chem., 289, 2014
|
|
4AQR
 
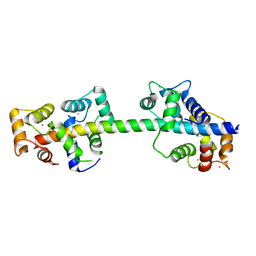 | | Crystal structure of calmodulin in complex with the regulatory domain of a plasma-membrane Ca2+-ATPase | | Descriptor: | CALCIUM ION, CALCIUM-TRANSPORTING ATPASE 8, PLASMA MEMBRANE-TYPE, ... | | Authors: | Tidow, H, Poulsen, L.R, Andreeva, A, Hein, K.L, Palmgren, M.G, Nissen, P. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Bimodular Mechanism of Calcium Control in Eukaryotes
Nature, 491, 2012
|
|
459D
 
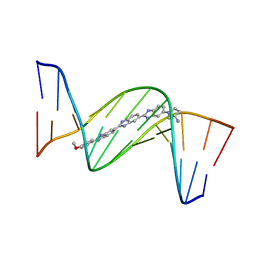 | | DNA MINOR-GROOVE RECOGNITION OF A TRIS-BENZIMIDAZOLE DRUG | | Descriptor: | 2''-(4-METHOXYPHENYL)-5-(3-AMINO-1-PYRROLIDINYL)-2,5',2',5''-TRI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*(CBR)P*AP*TP*AP*TP*TP*TP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*AP*TP*GP*CP*G)-3') | | Authors: | Aymami, J, Nunn, C.M, Neidle, S. | | Deposit date: | 1999-03-10 | | Release date: | 1999-06-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA minor groove recognition of a non-self-complementary AT-rich sequence by a tris-benzimidazole ligand.
Nucleic Acids Res., 27, 1999
|
|
1WET
 
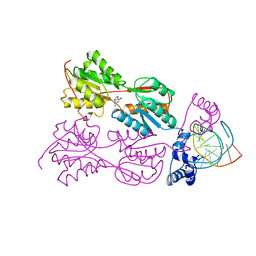 | | STRUCTURE OF THE PURR-GUANINE-PURF OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*TP*TP*TP*TP*CP*GP*T )-3'), GUANINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Glasfeld, A, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-04-27 | | Release date: | 1997-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the PurR-guanine-purF operator complex reveals the contributions of complementary electrostatic surfaces and a water-mediated hydrogen bond to corepressor specificity and binding affinity.
J.Biol.Chem., 272, 1997
|
|
3S74
 
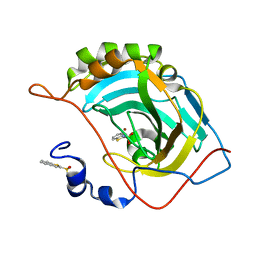 | |
1J8T
 
 | | Catalytic Domain of Human Phenylalanine Hydroxylase Fe(II) | | Descriptor: | FE (II) ION, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Andersen, O.A, Flatmark, T, Hough, E. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution crystal structures of the catalytic domain of human phenylalanine hydroxylase in its catalytically active Fe(II) form and binary complex with tetrahydrobiopterin.
J.Mol.Biol., 314, 2001
|
|
4ITZ
 
 | |
4OQ1
 
 | |
3S75
 
 | |
3S76
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1H-imidazole-2-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3S73
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4A6Y
 
 | |
3UBW
 
 | | Complex of 14-3-3 isoform epsilon, a Mlf1 phosphopeptide and a small fragment hit from a FBDD screen | | Descriptor: | (3S)-pyrrolidin-3-ol, 14-3-3 protein epsilon, Myeloid leukemia factor 1, ... | | Authors: | Molzan, M, Weyand, M, Rose, R, Ottmann, C. | | Deposit date: | 2011-10-25 | | Release date: | 2012-01-25 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights of the MLF1/14-3-3 interaction.
Febs J., 279, 2012
|
|
1ARB
 
 | |
1OJQ
 
 | | The crystal structure of C3stau2 from S. aureus | | Descriptor: | ADP-RIBOSYLTRANSFERASE | | Authors: | Evans, H.R, Sutton, J.M, Holloway, D.E, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2003-07-15 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Crystal Structure of C3Stau2 from Staphylococcus Aureus and its Complex with Nad
J.Biol.Chem., 278, 2003
|
|
