4BY0
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-3,3'- difluoro-(1,1'-biphenyl)-4-carboxamide | | Descriptor: | (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-3,3'-difluoro-(1,1'-biphenyl)-4-carboxamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Choi, J.Y, Calvet, C.M, Vierira, D.F, Gunatilleke, S.S, Cameron, M.D, McKerrow, J.H, Podust, L.M, Roush, W.R. | | Deposit date: | 2013-07-16 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | R-Configuration of 4-Aminopyridyl-Based Inhibitors of Cyp51 Confers Superior Efficacy Against Trypanosoma Cruzi
Acs Med.Chem.Lett., 5, 2014
|
|
7GPM
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z979742720 | | Descriptor: | 5-(propan-2-yl)-3-[(2S)-pyrrolidin-2-yl]-1,2,4-oxadiazole, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GPD
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z57472297 | | Descriptor: | 1-[2-methyl-1,3-bis(oxidanyl)propan-2-yl]-3-phenyl-urea, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SX5
 
 | |
7H2K
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z18769001 | | Descriptor: | (2S)-2-(2-cyanophenoxy)propanamide, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
5SX4
 
 | | Crystal Structure of panitumumab in complex with epidermal growth factor receptor domain 3. | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Sickmier, E.A, Kurzeja, R.J.M, Michelsen, K, Mukta, V, Yang, E, Tasker, A.S. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Panitumumab EGFR Complex Reveals a Binding Mechanism That Overcomes Cetuximab Induced Resistance.
Plos One, 11, 2016
|
|
7GPC
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z57328552 | | Descriptor: | 2-methyl-2-{[(3-methylthiophen-2-yl)methyl]amino}propan-1-ol, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q20
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000524a | | Descriptor: | 1-[2-methyl-1,3-bis(oxidanyl)propan-2-yl]-3-phenyl-urea, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FVN
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z371875396 | | Descriptor: | N-(propan-2-yl)-4-(thiophene-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FV7
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z1929967066 | | Descriptor: | (3R)-4-(furan-2-carbonyl)-3-methyl-N-(propan-2-yl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7G9L
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z111782404 | | Descriptor: | (2S)-2-[([1,1'-biphenyl]-4-yl)oxy]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7P6S
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2, pentane-1,5-diol | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6R
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form I) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OZX
 
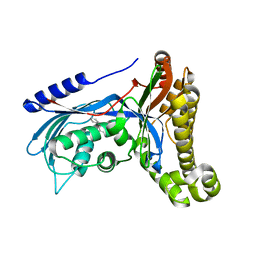 | | Structure of human galactokinase 1 bound with azepan-1-yl(2,6-difluorophenyl)methanone | | Descriptor: | (azepan-1-yl)(2,6-difluorophenyl)methanone, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human galactokinase 1 bound with azepan-1-yl(2,6-difluorophenyl)methanone
To Be Published
|
|
6OOD
 
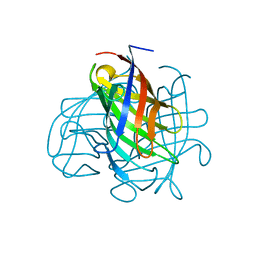 | | Structure of the pterocarpan synthase dirigent protein PsPTS1 | | Descriptor: | pterocarpan synthase dirigent protein PsPTS1 | | Authors: | Smith, C.A. | | Deposit date: | 2019-04-23 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pterocarpan synthase (PTS) structures suggest a common quinone methide-stabilizing function in dirigent proteins and proteins with dirigent-like domains.
J.Biol.Chem., 295, 2020
|
|
5Q1R
 
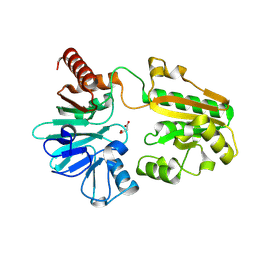 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000291a | | Descriptor: | 3-(azepan-1-ylmethyl)-1~{H}-indole, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5S9S
 
 | |
5SA2
 
 | |
5S9X
 
 | | PanDDA analysis group deposition -- Crystal Structure of Trypanosoma brucei Trypanothione reductase in complex with Z1899842917 | | Descriptor: | 3-[(3,5-dimethyl-1,2-oxazol-4-yl)methyl]-5-methyl-1,3,4-thiadiazol-2(3H)-one, BROMIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fiorillo, A, Ilari, A. | | Deposit date: | 2021-05-18 | | Release date: | 2022-06-15 | | Last modified: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Innovative Approach for a Classic Target: Fragment Screening on Trypanothione Reductase Reveals New Opportunities for Drug Design.
Front Mol Biosci, 9, 2022
|
|
5S9W
 
 | |
5SA1
 
 | |
5S9T
 
 | |
5S9Z
 
 | |
5S9Y
 
 | |
6PU2
 
 | | Dark, Mutant H275T , 100K, PCM Myxobacterial Phytochrome, P2 | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
