5XJ4
 
 | | Complex structure of durvalumab-scFv/PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1, durvalumab-VH, durvalumab-VL | | Authors: | Tan, S, Liu, K, Chai, Y, Gao, G.F, Qi, J. | | Deposit date: | 2017-04-29 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct PD-L1 binding characteristics of therapeutic monoclonal antibody durvalumab
Protein Cell, 9, 2018
|
|
5XPW
 
 | | Structure of amphioxus IgVJ-C2 molecule | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
2JJT
 
 | | Structure of human CD47 in complex with human signal regulatory protein (SIRP) alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUKOCYTE SURFACE ANTIGEN CD47, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE SUBSTRATE 1 | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47.
Mol. Cell, 31, 2008
|
|
5VST
 
 | | Crystal structure of murine CEACAM1b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Biliary glycoprotein | | Authors: | Peng, G, Yang, Y, Pasquarella, J.R, Xu, L, Qian, Z, Holmes, K.V, Li, F. | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism for coronavirus-driven evolution of mouse receptor
J. Biol. Chem., 292, 2017
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2JJS
 
 | | Structure of human CD47 in complex with human signal regulatory protein (SIRP) alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, LEUKOCYTE SURFACE ANTIGEN CD47, ... | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47.
Mol. Cell, 31, 2008
|
|
2GHW
 
 | | Crystal structure of SARS spike protein receptor binding domain in complex with a neutralizing antibody, 80R | | Descriptor: | CHLORIDE ION, Spike glycoprotein, anti-sars scFv antibody, ... | | Authors: | Hwang, W.C, Lin, Y, Santelli, E, Sui, J, Jaroszewski, L, Stec, B, Farzan, M, Marasco, W.A, Liddington, R.C. | | Deposit date: | 2006-03-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
J.Biol.Chem., 281, 2006
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6AW3
 
 | | Crystal structure of the HopQ-CEACAM3 L44Q complex | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 3, HopQ | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
6ARQ
 
 | | Crystal structure of CD96 (D1) bound to CD155/necl-5 (D1-3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor, T-cell surface protein tactile, ... | | Authors: | Deuss, F.A, Watson, G.M, Rossjohn, J, Berry, R. | | Deposit date: | 2017-08-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Basis for CD96 Immune Receptor Recognition of Nectin-like Protein-5, CD155.
Structure, 27, 2019
|
|
6C6Q
 
 | |
6AW2
 
 | | Crystal structure of the HopQ-CEACAM1 complex | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, HopQ | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
6AVZ
 
 | | Crystal structure of the HopQ-CEACAM3 WT complex | | Descriptor: | CALCIUM ION, Carcinoembryonic antigen-related cell adhesion molecule 3, HopQ, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
2PTT
 
 | |
2PET
 
 | | Lutheran glycoprotein, N-terminal domains 1 and 2. | | Descriptor: | Lutheran blood group glycoprotein | | Authors: | Burton, N, Brady, R.L. | | Deposit date: | 2007-04-03 | | Release date: | 2007-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Laminin 511/521-binding site on the Lutheran blood group glycoprotein is located at the flexible junction of Ig domains 2 and 3.
Blood, 110, 2007
|
|
2PF6
 
 | | Lutheran glycoprotein, N-terminal domains 1 and 2 | | Descriptor: | Lutheran blood group glycoprotein | | Authors: | Burton, N, Brady, R.L. | | Deposit date: | 2007-04-04 | | Release date: | 2007-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Laminin 511/521-binding site on the Lutheran blood group glycoprotein is located at the flexible junction of Ig domains 2 and 3.
Blood, 110, 2007
|
|
2OZ4
 
 | | Structural Plasticity in IgSF Domain 4 of ICAM-1 Mediates Cell Surface Dimerization | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB FRAGMENT LIGHT CHAIN, ... | | Authors: | Chen, X, Kim, T.D, Carman, C.V, Mi, L, Song, G, Springer, T.A. | | Deposit date: | 2007-02-23 | | Release date: | 2007-10-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity in Ig superfamily domain 4 of ICAM-1 mediates cell surface dimerization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1L6Z
 
 | | CRYSTAL STRUCTURE OF MURINE CEACAM1A[1,4]: A CORONAVIRUS RECEPTOR AND CELL ADHESION MOLECULE IN THE CEA FAMILY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, biliary glycoprotein C | | Authors: | Tan, K, Zelus, B.D, Meijers, R, Liu, J.-H, Bergelson, J.M, Duke, N, Zhang, R, Joachimiak, A, Holmes, K.V, Wang, J.-H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | CRYSTAL STRUCTURE OF MURINE sCEACAM1a[1,4]: A CORONAVIRUS RECEPTOR IN THE CEA FAMILY
Embo J., 21, 2002
|
|
1NBQ
 
 | | Crystal Structure of Human Junctional Adhesion Molecule Type 1 | | Descriptor: | Junctional adhesion molecule 1 | | Authors: | Prota, A.E, Campbell, J.A, Schelling, P, Forrest, J.C, Watson, M.J, Peters, T.R, Aurrand-Lions, M, Imhof, B.A, Dermody, T.S, Stehle, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-04-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human junctional adhesion molecule 1: Implications for reovirus binding
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1SQ2
 
 | | Crystal Structure Analysis of the Nurse Shark New Antigen Receptor (NAR) Variable Domain in Complex With Lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Stanfield, R.L, Dooley, H, Flajnik, M.F, Wilson, I.A. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a shark single-domain antibody V region in complex with lysozyme.
Science, 305, 2004
|
|
1J1O
 
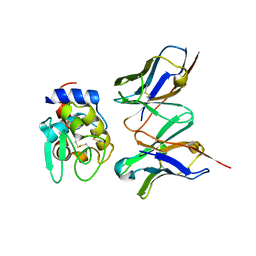 | | Crystal Structure of HyHEL-10 Fv mutant LY50F complexed with hen egg white lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Yokota, A, Tsumoto, K, Shiroishi, M, Kondo, H, Kumagai, I. | | Deposit date: | 2002-12-13 | | Release date: | 2003-01-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Role of Hydrogen Bonding via Interfacial Water Molecules in Antigen-Antibody Complexation. THE HyHEL-10-HEL INTERACTION
J.BIOL.CHEM., 278, 2003
|
|
1J1X
 
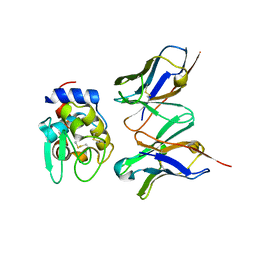 | | Crystal Structure of HyHEL-10 Fv mutant LS93A complexed with hen egg white lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Yokota, A, Tsumoto, K, Shiroishi, M, Kondo, H, Kumagai, I. | | Deposit date: | 2002-12-20 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Role of Hydrogen Bonding via Interfacial Water Molecules in Antigen-Antibody Complexation. THE HyHEL-10-HEL INTERACTION
J.BIOL.CHEM., 278, 2003
|
|
7BPK
 
 | | Zika virus envelope protein mutant bound to mAb | | Descriptor: | Envelope protein, IG c307_light_IGLV1-51_IGLJ2, Z3L1 Heavy chain | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protective Zika vaccines engineered to eliminate enhancement of dengue infection via immunodominance switch.
Nat.Immunol., 22, 2021
|
|
1S78
 
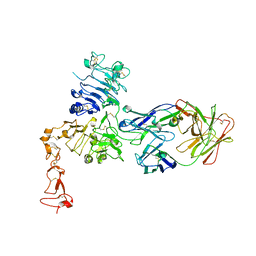 | | Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, ... | | Authors: | Franklin, M.C, Carey, K.D, Vajdos, F.F, Leahy, D.J, de Vos, A.M, Sliwkowski, M.X. | | Deposit date: | 2004-01-29 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex.
Cancer Cell, 5, 2004
|
|
3CFK
 
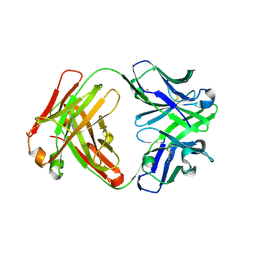 | | Crystal structure of catalytic elimination antibody 34E4, triclinic crystal form | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, CATALYTIC ANTIBODY FAB 34E4 HEAVY CHAIN,Uncharacterized protein, ... | | Authors: | Debler, E.W, Wilson, I.A. | | Deposit date: | 2008-03-04 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational isomerism can limit antibody catalysis.
J.Biol.Chem., 283, 2008
|
|
