5PTY
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 176) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PUD
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 191) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PUT
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 207) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2PS7
 
 | | Y295F trichodiene synthase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Trichodiene synthase | | Authors: | Vedula, L.S, Cane, D.E, Christianson, D.W. | | Deposit date: | 2007-05-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and mechanistic analysis of trichodiene synthase using site-directed mutagenesis: probing the catalytic function of tyrosine-295 and the asparagine-225/serine-229/glutamate-233-Mg2+B motif.
Arch.Biochem.Biophys., 469, 2008
|
|
7LMO
 
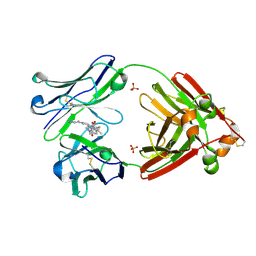 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 34 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-7-(1H-imidazole-5-carbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione] | | Descriptor: | (5~{R})-3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-7-(1~{H}-imidazol-4-ylcarbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
3LMJ
 
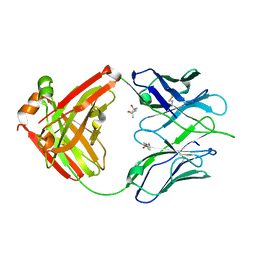 | | Structure of human anti HIV 21c Fab | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Heavy chain of anti HIV Fab from human 21c antibody, Light chain of anti HIV Fab from human 21c antibody | | Authors: | Diskin, R, Marcovecchio, P.M, Bjorkman, P.J. | | Deposit date: | 2010-01-30 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5PV9
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 223) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PVK
 
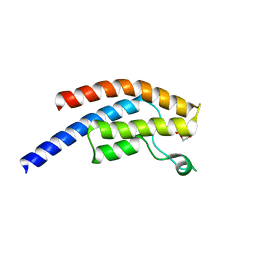 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 235) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PW1
 
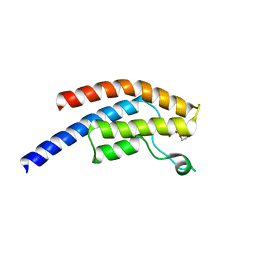 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 252) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PWG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 3) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PWW
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 20) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4Q4P
 
 | | tRNA-Guanine Transglycosylase (TGT) in Complex with 2-{[2-(PIPERIDIN-1-YL)ETHYL]AMINO}-3,5-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE | | Descriptor: | 2-{[2-(piperidin-1-yl)ethyl]amino}-3,5-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Beyond Affinity: Enthalpy-Entropy Factorization Unravels Complexity of a Flat Structure-Activity Relationship for Inhibition of a tRNA-Modifying Enzyme.
J.Med.Chem., 57, 2014
|
|
5PXC
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 36) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PXS
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 52) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2VOT
 
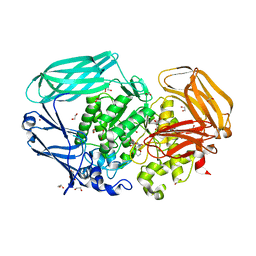 | | Structural and biochemical evidence for a boat-like transition state in beta-mannosidases | | Descriptor: | (5R,6R,7S,8R)-6,7,8-trihydroxy-5-(hydroxymethyl)-2-[(phenylamino)methyl]-5,6,7,8-tetrahydro-1H-imidazo[1,2-a]pyridin-4-ium, 1,2-ETHANEDIOL, BETA-MANNOSIDASE, ... | | Authors: | Tailford, L.N, Offen, W.A, Smith, N, Dumon, C, Moreland, C, Gratien, J, Heck, M.-P, Stick, R.V, Bleriot, Y, Vasella, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2008-02-20 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biochemical Evidence for a Boat-Like Transition State in Beta-Mannosidases.
Nat.Chem.Biol., 4, 2008
|
|
4M6R
 
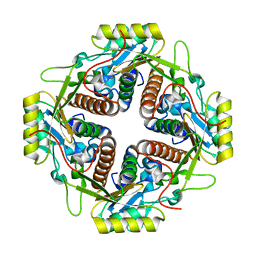 | | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme | | Descriptor: | Methylthioribulose-1-phosphate dehydratase, ZINC ION | | Authors: | Kang, W, Hong, S.H, Lee, H.M, Kim, N.Y, Lim, Y.C, Le, L.T.M, Lim, B, Kim, H.C, Kim, T.Y, Ashida, H, Yokota, A, Hah, S.S, Chun, K.H, Jung, Y.K, Yang, J.K. | | Deposit date: | 2013-08-10 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5PY6
 
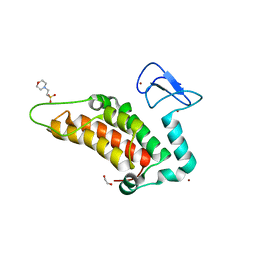 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 66) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4QGX
 
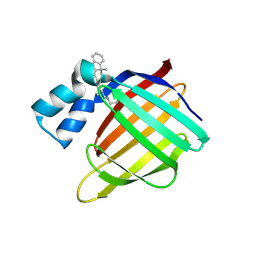 | | Crystal structure of the R132K:R111L:L121E mutant of Cellular Retinoic Acid Binding ProteinII complexed with a synthetic ligand (Merocyanine) at 1.47 angstrom resolution | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, Cellular retinoic acid-binding protein 2 | | Authors: | Nosrati, M, Yapici, I, Geiger, J.H. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
5PZ1
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 97) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3CVV
 
 | | Drosophila melanogaster (6-4) photolyase bound to ds DNA with a T-T (6-4) photolesion and F0 cofactor | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), ... | | Authors: | Glas, A.F, Maul, M.J, Cryle, M.J, Barends, T.R.M, Schneider, S, Kaya, E, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The archaeal cofactor F0 is a light-harvesting antenna chromophore in eukaryotes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5PZI
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 114) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3LOP
 
 | | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum | | Descriptor: | 1,2-ETHANEDIOL, LEUCINE, MAGNESIUM ION, ... | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum
To be Published
|
|
5Q0M
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 5-{[(3beta,5beta,14beta,17alpha)-3-hydroxy-24-oxocholan-24-yl]amino}benzene-1,3-dicarboxylic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0W
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-({5-bromo-1'-[(2-chlorophenyl)sulfonyl]-2-oxospiro[indole-3,4'-piperidin]-1(2H)-yl}methyl)benzoic acid, Bile acid receptor, cDNA FLJ76652, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
4QKM
 
 | | Influenza A M2 wild type TM domain at low pH in the lipidic cubic phase under room temperature diffraction conditions | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Thomaston, J.L, DeGrado, W.F. | | Deposit date: | 2014-06-06 | | Release date: | 2015-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | High-resolution structures of the M2 channel from influenza A virus reveal dynamic pathways for proton stabilization and transduction.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
