1GW1
 
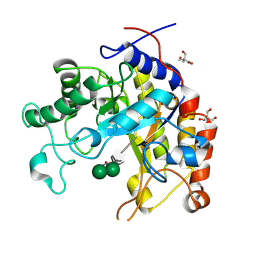 | | Substrate distortion by beta-mannanase from Pseudomonas cellulosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DINITROPHENYLENE, MANNAN ENDO-1,4-BETA-MANNOSIDASE, ... | | Authors: | Ducros, V, Zechel, D.L, Gilbert, H.J, Szabo, L, Withers, S.G, Davies, G.J. | | Deposit date: | 2002-03-01 | | Release date: | 2002-09-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Distortion by a Beta-Mannanase: Snapshots of the Michaelis and Covalent-Intermediate Complexes Suggest a B2,5 Conformation for the Transition State
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
5SLN
 
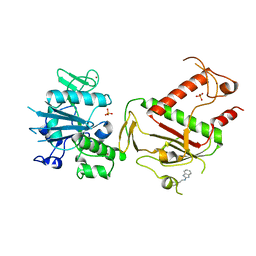 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z57299529 | | Descriptor: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4AYG
 
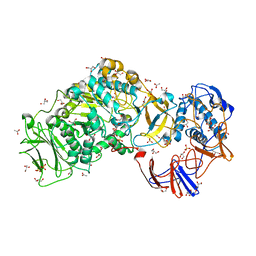 | | Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in orthorhombic apo-form | | Descriptor: | ACETIC ACID, CALCIUM ION, GLUCANSUCRASE, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2012-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility of Truncated and Full-Length Glucansucrase Gtf180 Enzymes from Lactobacillus Reuteri 180.
FEBS J., 281, 2014
|
|
7UGM
 
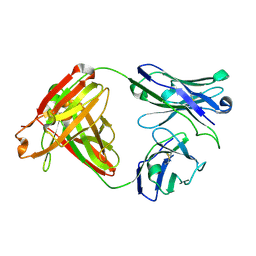 | |
3F3V
 
 | | Kinase domain of cSrc in complex with inhibitor RL45 (Type II) | | Descriptor: | 1-{4-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Grutter, C, Kluter, S, Getlik, M, Rauh, D. | | Deposit date: | 2008-10-31 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hybrid compound design to overcome the gatekeeper T338M mutation in cSrc
J.Med.Chem., 52, 2009
|
|
5SKZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z57258487 | | Descriptor: | N-{[4-(dimethylamino)phenyl]methyl}-4H-1,2,4-triazol-4-amine, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6ELE
 
 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, fAB heavy chain, ... | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
5SL2
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z100643660 | | Descriptor: | N,1-dimethyl-1H-indole-3-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q25
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000532a | | Descriptor: | 1~{H}-indol-6-yl-(4-methylpiperazin-1-yl)methanone, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SMA
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434890 | | Descriptor: | 4-methyl-N-phenylpiperazine-1-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2W1A
 
 | | Non-covalent complex between dahp synthase and chorismate mutase from Mycobacterium tuberculosis with bound tsa | | Descriptor: | 3-DEOXY-D-ARABINO-HEPTULOSONATE 7-PHOSPHATE SYNTHASE AROG, 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHORISMATE MUTASE, ... | | Authors: | Okvist, M, Sasso, S, Roderer, K, Gamper, M, Codoni, G, Krengel, U, Kast, P. | | Deposit date: | 2008-10-16 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Function of a Complex between Chorismate Mutase and Dahp Synthase: Efficiency Boost for the Junior Partner.
Embo J., 28, 2009
|
|
4QBM
 
 | | Crystal structure of human BAZ2A bromodomain in complex with a diacetylated histone 4 peptide (H4K16acK20ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2A, histone H4 peptide with sequence Gly-Ala-Lys(ac)-Arg-His-Arg-Lys(ac)-Val-Leu | | Authors: | Tallant, C, Nunez-Alonso, G, Picaud, S, Filippakopoulos, P, Krojer, T, Williams, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
4II4
 
 | |
3F8J
 
 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group C222(1) | | Descriptor: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1, ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-12 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
3F97
 
 | | Crystal structure of human plasma platelet activating factor acetylhydrolase covalently inhibited by soman | | Descriptor: | (1R)-1,2,2-TRIMETHYLPROPYL (R)-METHYLPHOSPHINATE, ACETATE ION, Platelet-activating factor acetylhydrolase, ... | | Authors: | Samanta, U, Bahnson, B.J. | | Deposit date: | 2008-11-13 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of human group-VIIA phospholipase A2 inhibited by organophosphorus nerve agents exhibit non-aged complexes.
Biochem Pharmacol, 78, 2009
|
|
5SAV
 
 | | DDR1, N-[2-[3-(2-aminopyrimidin-5-yl)oxyphenyl]ethyl]-3-(trifluoromethoxy)benzamide, 1.760A, P212121, Rfree=23.5% | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-(2-{3-[(2-aminopyrimidin-5-yl)oxy]phenyl}ethyl)-3-(trifluoromethoxy)benzamide | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
4AVB
 
 | | Crystal structure of protein lysine acetyltransferase Rv0998 in complex with acetyl CoA and cAMP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7HUL
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0031733-001 (A71EV2A-x3181) | | Descriptor: | 1-methyl-N-[2-(1H-1,2,4-triazol-1-yl)ethyl]-1H-indazole-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
5SAZ
 
 | | DDR1, 3-chloro-N-[4-chloro-3-(1H-pyrrolo[2,3-b]pyridin-5-ylcarbamoyl)phenyl]-4-(2-hydroxyethylamino)benzamide, 1.802A, P212121, Rfree=22.2% | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-5-{3-chloro-4-[(2-hydroxyethyl)amino]benzamido}-N-(1H-pyrrolo[2,3-b]pyridin-5-yl)benzamide, CHLORIDE ION, ... | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
3PUK
 
 | | Re-refinement of the crystal structure of Munc18-3 and Syntaxin4 N-peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 3 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.054 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5SAU
 
 | | DDR1, 3-[2-(6-aminopyridin-3-yl)ethynyl]-N-[3-(trifluoromethyl)phenyl]benzamide, 1.800A, P212121, Rfree=23.1% | | Descriptor: | 3-[(6-aminopyridin-3-yl)ethynyl]-N-[3-(trifluoromethyl)phenyl]benzamide, Epithelial discoidin domain-containing receptor 1, MALONATE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
5SAX
 
 | | DDR1, 2-[3-(2-pyridin-3-ylethynyl)phenyl]-N-[3-(trifluoromethyl)phenyl]acetamide, 1.902A, second P212121 form, Rfree=25.4%, second form | | Descriptor: | 2-{3-[(pyridin-3-yl)ethynyl]phenyl}-N-[3-(trifluoromethyl)phenyl]acetamide, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
5SB0
 
 | | DDR1, N-[[2-(2-pyridin-3-yloxyethyl)cyclohexyl]methyl]-3-(trifluoromethoxy)benzamide, 1.970A, P212121, Rfree=25.6% | | Descriptor: | CHLORIDE ION, Epithelial discoidin domain-containing receptor 1, IODIDE ION, ... | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
4AVP
 
 | | Crystal structure of the DNA-binding domain of human ETV1. | | Descriptor: | 1,2-ETHANEDIOL, ETS TRANSLOCATION VARIANT 1 | | Authors: | Allerston, C.K, Cooper, C.D.O, Krojer, T, Chaikuad, A, Filippakopoulos, P, Canning, P, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2012-05-28 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
5SAW
 
 | | DDR1, 2-[3-(2-pyridin-3-ylethynyl)phenyl]-N-[3-(trifluoromethyl)phenyl]acetamide, 1.601A, P212121, Rfree=22.6% | | Descriptor: | 2-{3-[(pyridin-3-yl)ethynyl]phenyl}-N-[3-(trifluoromethyl)phenyl]acetamide, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
