5SR9
 
 | |
7GIR
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-971238d3-1 (Mpro-P0125) | | Descriptor: | (4S)-6-chloro-4-hydroxy-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5SRC
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5198562500 - (R,R) and (R,S) isomers | | Descriptor: | (2R)-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl](hydroxy)acetic acid, (2S)-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl](hydroxy)acetic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4TVG
 
 | | HIV Protease (PR) dimer in closed form with pepstatin in active site and fragment AK-2097 in the outside/top of flap | | Descriptor: | 3-(morpholin-4-ylmethyl)-1H-indole-6-carboxylic acid, GLYCEROL, HIV Protease, ... | | Authors: | Tiefenbrunn, T, Kislukhin, A, Finn, M.G, Stout, C.D. | | Deposit date: | 2014-06-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Distinguishing Binders from False Positives by Free Energy Calculations: Fragment Screening Against the Flap Site of HIV Protease.
J.Phys.Chem.B, 119, 2015
|
|
5SSN
 
 | |
5SOO
 
 | |
5SOM
 
 | |
5SSM
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5459166256 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-[(1H-benzimidazole-5-carbonyl)amino]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-[(1H-benzimidazole-5-carbonyl)amino]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2YY0
 
 | | Crystal Structure of MS0802, c-Myc-1 binding protein domain from Homo sapiens | | Descriptor: | C-Myc-binding protein | | Authors: | Xie, Y, Wang, H, Ihsanawati, K.T, Kishishita, S, Takemoto, C, Shirozu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2008-04-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | crystal structure of c-Myc-1 binding protein domain from Homo sapiens
To be Published
|
|
7GJL
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-UNI-3735e77e-1 (Mpro-P0600) | | Descriptor: | (3R)-3-(3,4-dichlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5SOK
 
 | |
5SOJ
 
 | |
5SOQ
 
 | |
5SOV
 
 | |
5SON
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000920153280 - (R) isomer | | Descriptor: | 3-{[(2R)-2-phenylpropyl]sulfanyl}-7H-[1,2,4]triazolo[4,3-b][1,2,4]triazole, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPA
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894417 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-(4-carbamamidobenzamido)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-(4-carbamamidobenzamido)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPB
 
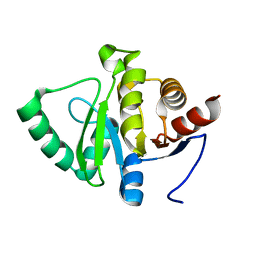 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894404 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-[4-(phenylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[4-(phenylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2C9Y
 
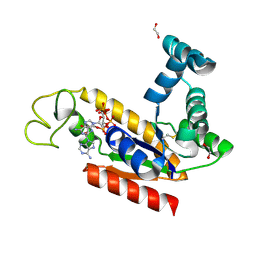 | | Structure of human adenylate kinase 2 | | Descriptor: | 1,2-ETHANEDIOL, ADENYLATE KINASE ISOENZYME 2, MITOCHONDRIAL, ... | | Authors: | Bunkoczi, G, Filippakopoulos, P, Debreczeni, J.E, Turnbull, A, Papagrigoriou, E, Savitsky, P, Colebrook, S, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Human Adenylate Kinase 2
To be Published
|
|
5SOY
 
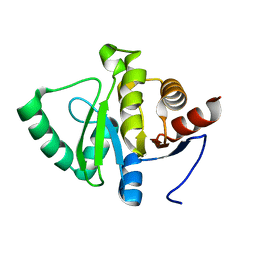 | |
3EQG
 
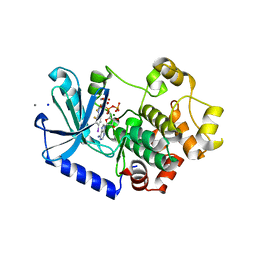 | |
4QAM
 
 | | Crystal Structure of the RPGR RCC1-like domain in complex with the RPGR-interacting domain of RPGRIP1 | | Descriptor: | GLYCEROL, MAGNESIUM ION, X-linked retinitis pigmentosa GTPase regulator, ... | | Authors: | Remans, K, Buerger, M, Vetter, I.R, Wittinghofer, A. | | Deposit date: | 2014-05-05 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | C2 domains as protein-protein interaction modules in the ciliary transition zone.
Cell Rep, 8, 2014
|
|
5SFE
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c2(nc1nc(cn1cc2)c3cc(ccc3)F)NC(c4c(cnn4C)C(N(C)C)=O)=O, micromolar IC50=0.011191 | | Descriptor: | MAGNESIUM ION, N~5~-[(2P,4S)-2-(3-fluorophenyl)imidazo[1,2-a]pyrimidin-7-yl]-N~4~,N~4~,1-trimethyl-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Gobbi, L, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SP0
 
 | |
5SP9
 
 | |
5SP7
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010903509 - (S,S) isomer | | Descriptor: | (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-1,2,3,4-tetrahydronaphthalene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
