4EZE
 
 | | Crystal structure of had family hydrolase t0658 from Salmonella enterica subsp. enterica serovar Typhi (Target EFI-501419) | | Descriptor: | CHLORIDE ION, Haloacid dehalogenase-like hydrolase, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-02 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of had hydrolase t0658 from Salmonella enterica (Target EFI-501419)
To be Published
|
|
4F7A
 
 | |
3H7K
 
 | |
2NNK
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
3H0N
 
 | |
3H12
 
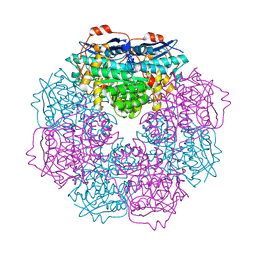 | | Crystal structure of putative mandelate racemase from Bordetella Bronchiseptica RB50 | | Descriptor: | SODIUM ION, mandelate racemase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative mandelate racemase from Bordetella Bronchiseptica RB50
To be Published
|
|
1TV9
 
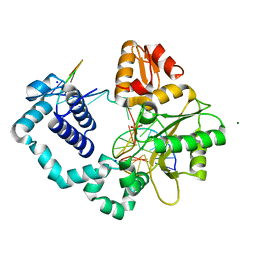 | | HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA CONTAINING A MISMATCHED TEMPLATE ADENINE AND INCOMING CYTIDINE | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Krahn, J.M, Beard, W.M, Wilson, S.H. | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | STRUCTURAL INSIGHTS INTO DNA POLYMERASE BETA DETERRENTS FOR MISINCORPORATION SUPPORT OF AN INDUCED-FIT MECHANISM FOR FIDELITY
Structure, 12, 2004
|
|
4FDZ
 
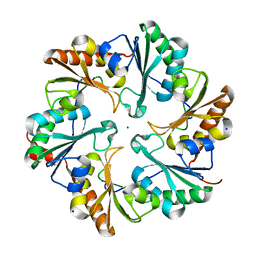 | | EutL from Clostridium perfringens, Crystallized Under Reducing Conditions | | Descriptor: | Ethanolamine utilization protein, SODIUM ION | | Authors: | Thompson, M.C, Cascio, D, Crowley, C.S, Kopstein, J.S, Yeates, T.O. | | Deposit date: | 2012-05-29 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein.
Protein Sci., 24, 2015
|
|
4F5Q
 
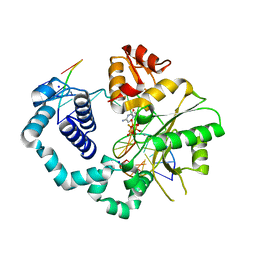 | | Closed ternary complex of R283K DNA polymerase beta | | Descriptor: | 2'-deoxy-5'-O-[(S)-{difluoro[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}(hydroxy)phosphoryl]cytidine, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Beard, W.A, Wilson, S.H. | | Deposit date: | 2012-05-13 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of dNTP Intermediate States during DNA Polymerase Active Site Assembly.
Structure, 20, 2012
|
|
4FAV
 
 | |
3GRD
 
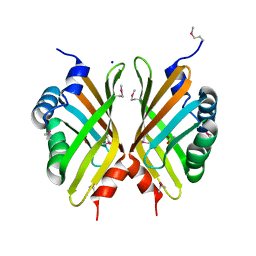 | |
3GSH
 
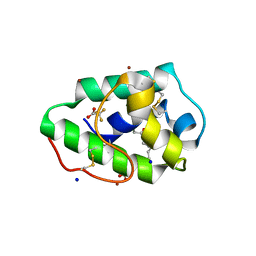 | | Three-dimensional structure of a post translational modified barley LTP1 | | Descriptor: | (12E)-10-oxooctadec-12-enoic acid, Non-specific lipid-transfer protein 1, SODIUM ION, ... | | Authors: | Lascombe, M.B, Prange, T, Bakan, B, Marion, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-12-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of oxylipin-conjugated barley LTP1 highlights the unique plasticity of the hydrophobic cavity of these plant lipid-binding proteins.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
3GNK
 
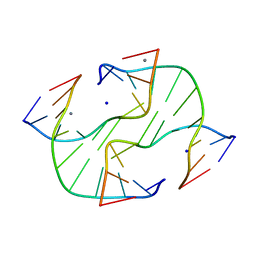 | |
2QFP
 
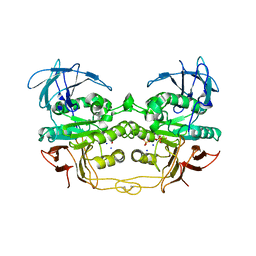 | | Crystal structure of red kidney bean purple acid phosphatase in complex with fluoride | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, FLUORIDE ION, ... | | Authors: | Guddat, L.W, Schenk, G.S, Gahan, L.R, Elliot, T.W, Leung, E. | | Deposit date: | 2007-06-27 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a purple acid phosphatase, representing different steps of this enzyme's catalytic cycle.
Bmc Struct.Biol., 8, 2008
|
|
2NQL
 
 | | Crystal structure of a member of the enolase superfamily from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, Isomerase/lactonizing enzyme, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Enolase from Agrobacterium Tumefaciens C58
To be Published
|
|
2E54
 
 | |
2E7U
 
 | | Crystal structure of glutamate-1-semialdehyde 2,1-aminomutase from Thermus thermophilus HB8 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLYCEROL, Glutamate-1-semialdehyde 2,1-aminomutase, ... | | Authors: | Mizutani, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-14 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glutamate-1-semialdehyde 2,1-aminomutase from Thermus thermophilus HB8
To be Published
|
|
2QK9
 
 | | Human RNase H catalytic domain mutant D210N in complex with 18-mer RNA/DNA hybrid | | Descriptor: | 5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*GP*CP*AP*CP*T)-3', 5'-R(*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3', CITRATE ANION, ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Ghirlando, R, Cerritelli, S.M, Crouch, R.J, Yang, W. | | Deposit date: | 2007-07-10 | | Release date: | 2007-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription
Mol.Cell, 28, 2007
|
|
2R37
 
 | | Crystal structure of human glutathione peroxidase 3 (selenocysteine to glycine mutant) | | Descriptor: | CHLORIDE ION, Glutathione peroxidase 3, SODIUM ION | | Authors: | Pilka, E.S, Guo, K, Gileadi, O, Rojkowa, A, von Delft, F, Pike, A.C.W, Kavanagh, K.L, Johannson, C, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-29 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human glutathione peroxidase 3 (selenocysteine to glycine mutant).
TO BE PUBLISHED
|
|
6WOX
 
 | |
3V3R
 
 | | Crystal Structure of GES-11 | | Descriptor: | Extended spectrum class A beta-lactamase GES-11, IODIDE ION, SODIUM ION | | Authors: | Delbruck, H, Hoffmann, K.M.V, Bebrone, C. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Kinetic and crystallographic studies of extended-spectrum GES-11, GES-12, and GES-14 beta-lactamases.
Antimicrob.Agents Chemother., 56, 2012
|
|
6WSA
 
 | |
5X7G
 
 | | Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase | | Descriptor: | CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, GLYCEROL, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Suzuki, R, Momma, M, Funane, K. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isomaltooligosaccharide-binding structure ofPaenibacillussp. 598K cycloisomaltooligosaccharide glucanotransferase
Biosci. Rep., 37, 2017
|
|
6WU3
 
 | |
6WYF
 
 | |
