5CYS
 
 | |
1VM4
 
 | |
6YYZ
 
 | | tRNA-Guanine Transglycosylase (TGT) labeled with 5-fluorotryptophan in co-crystallized complex with 6-amino-2-(methylamino)-4-(2-((2R,3R,4R,5R)-3,4,5-trimethoxytetrahydrofuran-2-yl)ethyl)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-(methylamino)-4-(2-((2R,3R,4R,5R)-3,4,5-trimethoxytetrahydrofuran-2-yl)ethyl)-1H-imidazo[4,5-g]quinazolin-8(7H)-one, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Co-crystallization, 19F NMR and nanoESI-MS reveal dimer disturbing inhibitors and conformational changes at dimer contacts
To Be Published
|
|
9BFL
 
 | |
8Y0F
 
 | |
1VM2
 
 | |
1N2Y
 
 | | SOLUTION STRUCTURE OF SS-CYCLIZED CATESTATIN FRAGMENT FROM CHROMOGRANIN A | | Descriptor: | CATESTATIN | | Authors: | Preece, N.E, Nguyen, M, Mahata, M, Mahata, S.K, Mahapatra, N.R, Tsigelny, I, O'Connor, D.T. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Conformational preferences and activities of peptides from the catecholamine
release-inhibitory (catestatin) region of chromogranin A
Regul.Pept., 118, 2004
|
|
6QAX
 
 | | P31-43 | | Descriptor: | LEU-GLY-GLN-GLN-GLN-PRO-PHE-PRO-PRO-GLN-GLN-PRO-TYR | | Authors: | Calvanese, L, D'Auria, G, Falcigno, F. | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural insights on P31-43, a gliadin peptide able to promote an innate but not an adaptive response in celiac disease.
J.Pept.Sci., 25, 2019
|
|
8IM5
 
 | |
5VFK
 
 | |
1W4F
 
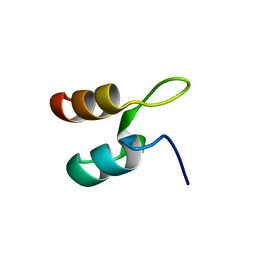 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
6QJB
 
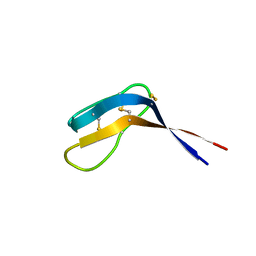 | | Truncated Evasin-3 (tEv3 17-56) | | Descriptor: | Evasin-3 | | Authors: | Denisov, S.S, Ippel, J.H, Heinzman, A.C.A, Koenen, R.R, Ortega-Gomez, A, Soehnlein, O, Hackeng, T.M, Dijkgraaf, I. | | Deposit date: | 2019-01-24 | | Release date: | 2019-07-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Tick saliva protein Evasin-3 modulates chemotaxis by disrupting CXCL8 interactions with glycosaminoglycans and CXCR2.
J.Biol.Chem., 294, 2019
|
|
1W4G
 
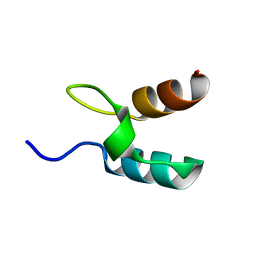 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state folding transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
8RPQ
 
 | |
8RQ6
 
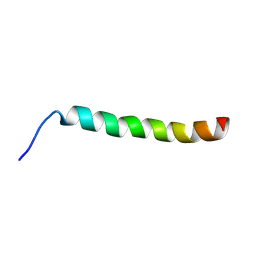 | |
5L06
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd Position | | Descriptor: | DNA (5'-D(*CP*GP*(5CM)P*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Miears, H.L, Hoppins, J.J, Gruber, D.R, Kasymov, R.D, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-07-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
7RC7
 
 | | Solution NMR Structure of [Ala19]Crp4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Conibear, A.C, Rosengren, K.J. | | Deposit date: | 2021-07-07 | | Release date: | 2021-08-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A conserved beta-bulge glycine residue facilitates folding and increases stability of the mouse alpha-defensin cryptdin-4
Peptide Science, 114, 2022
|
|
7LU8
 
 | |
7XRW
 
 | | Solution structure of T. brucei RAP1 | | Descriptor: | Repressor activator protein 1 | | Authors: | Yang, X, Pan, X.H, Ji, Z.Y, Wong, K.B, Zhang, M.J, Zhao, Y.X. | | Deposit date: | 2022-05-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The RRM-mediated RNA binding activity in T. brucei RAP1 is essential for VSG monoallelic expression.
Nat Commun, 14, 2023
|
|
5KQB
 
 | |
5KQC
 
 | |
1M3V
 
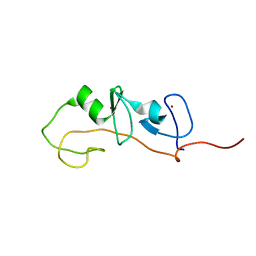 | | FLIN4: Fusion of the LIM binding domain of Ldb1 and the N-terminal LIM domain of LMO4 | | Descriptor: | ZINC ION, fusion of the LIM interacting domain of ldb1 and the N-terminal LIM domain of LMO4 | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H.Y, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2002-06-30 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
7LVE
 
 | |
8TNS
 
 | |
7LUW
 
 | |
