7Z8B
 
 | |
7Z7H
 
 | | Structure of P. luminescens TccC3-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, Actin, ... | | Authors: | Belyy, A, Raunser, S. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-29 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of threonine ADP-ribosylation of F-actin by a Tc toxin.
Nat Commun, 13, 2022
|
|
7ZDU
 
 | |
7ZIX
 
 | |
7ZDC
 
 | |
4PO5
 
 | | Crystal structure of allophycocyanin B from Synechocystis PCC 6803 | | Descriptor: | Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, PHYCOCYANOBILIN, ... | | Authors: | Pang, P.P, Dong, L.L, Sun, Y.F, Zeng, X.L, Ding, W.L, Scheer, H, Yang, X, Zhao, K.H. | | Deposit date: | 2014-02-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The structure of allophycocyanin B from Synechocystis PCC 6803 reveals the structural basis for the extreme redshift of the terminal emitter in phycobilisomes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7ZD5
 
 | | IF(apo/as isolated) conformation of CydDC (Dataset-1) | | Descriptor: | ATP-binding/permease protein CydC, ATP-binding/permease protein CydD | | Authors: | Wu, D, Safarian, S. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Dissecting the conformational complexity and mechanism of a bacterial heme transporter.
Nat.Chem.Biol., 19, 2023
|
|
7ZS0
 
 | |
7ZS1
 
 | |
7ZS2
 
 | |
7QKR
 
 | | Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris with Verapamil at 3.2 A resolution | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, Dexverapamil, MAGNESIUM ION, ... | | Authors: | Schleker, E.S.M, Reinhart, C. | | Deposit date: | 2021-12-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional investigation of ABC transporter STE6-2p from Pichia pastoris reveals unexpected interaction with sterol molecules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8AW5
 
 | | Cryo-EM structure of heme A synthase trimer from Aquifex aeolicus | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Heme O oxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hui, Z, Guoliang, Z. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of heme A synthase trimer from Aquifex aeolicus
To Be Published
|
|
8B4I
 
 | | Cryo-EM structure of the Neurospora crassa TOM core complex at 3.3 angstrom | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, DIUNDECYL PHOSPHATIDYL CHOLINE, Mitochondrial import receptor subunit Tom22, ... | | Authors: | Ornelas, P, Kuehlbrandt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Two conformations of the Tom20 preprotein receptor in the TOM holo complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7ZJV
 
 | |
7ZJU
 
 | |
7ZOX
 
 | | Nup93 in complex with xhNup93-Nb4i and xNup93-Nb2t | | Descriptor: | Nuclear pore complex protein Nup93, xNup93-Nb2t, xhNup93-Nb4i | | Authors: | Fu, Z, Guttler, T, Colom, M.S. | | Deposit date: | 2022-04-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A checkpoint function for Nup98 in nuclear pore formation suggested by novel inhibitory nanobodies.
Embo J., 2024
|
|
8AN1
 
 | | Structure of a first level Sierpinski triangle formed by a citrate synthase | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2022-08-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Emergence of fractal geometries in the evolution of a metabolic enzyme.
Nature, 628, 2024
|
|
8A9E
 
 | | Lysozyme, 9-11 fs FEL pulses as determined by XTCAV | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, GADOLINIUM ATOM, Lysozyme | | Authors: | Barends, T, Nass, K, Gorel, A, Schlichting, I. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Microcrystallization methods
To Be Published
|
|
8CUY
 
 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CV0
 
 | | KS-AT domains of mycobacterial Pks13 with outward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CUZ
 
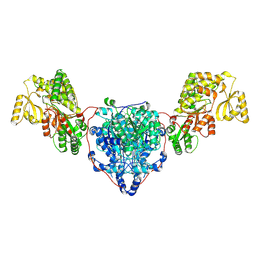 | | KS-AT domains of mycobacterial Pks13 with inward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A3O
 
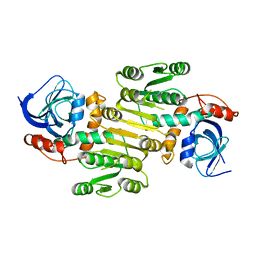 | | Structure of human Fy-4 | | Descriptor: | Quinone oxidoreductase-like protein 1 | | Authors: | Schuhmacher, J.S, Zerial, M. | | Deposit date: | 2022-06-08 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of mRNA binding by the human FERRY Rab5 effector complex.
Mol.Cell, 83, 2023
|
|
8CV1
 
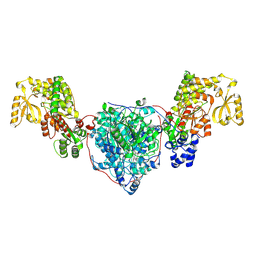 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A3P
 
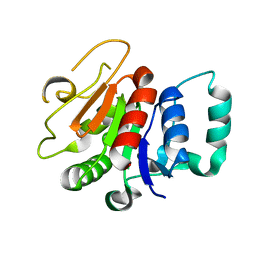 | | Structure of human Fy-5. | | Descriptor: | Glutamine amidotransferase-like class 1 domain-containing protein 1 | | Authors: | Schuhmacher, J.S, Zerial, M. | | Deposit date: | 2022-06-08 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of mRNA binding by the human FERRY Rab5 effector complex.
Mol.Cell, 83, 2023
|
|
7QP6
 
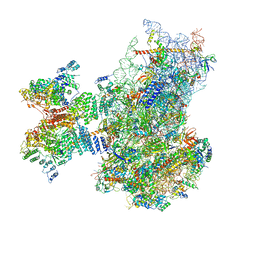 | | Structure of the human 48S initiation complex in open state (h48S AUG open) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
