9C55
 
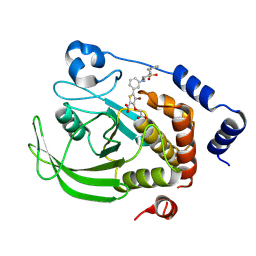 | | Crystal structure of human PTPN2 in complex with active site inhibitor | | Descriptor: | 5-(3-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}PHENYL)-4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Bester, S.M, Linwood, R, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-06-05 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Enhancing the apo protein tyrosine phosphatase non-receptor type 2 crystal soaking strategy through inhibitor-accessible binding sites.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
7MME
 
 | |
1H56
 
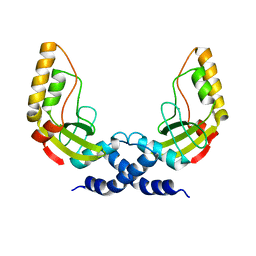 | | Structural and biochemical characterization of a new magnesium ion binding site near Tyr94 in the restriction endonuclease PvuII | | Descriptor: | MAGNESIUM ION, TYPE II RESTRICTION ENZYME PVUII | | Authors: | Spyrida, A, Matzen, C, Lanio, T, Jeltsch, A, Simoncsits, A, Athanasiadis, A, Scheuring-Vanamee, E, Kokkinidis, M, Pingoud, A. | | Deposit date: | 2001-05-20 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Biochemical Characterization of a New Mg(2+) Binding Site Near Tyr94 in the Restriction Endonuclease PvuII.
J.Mol.Biol., 331, 2003
|
|
2X87
 
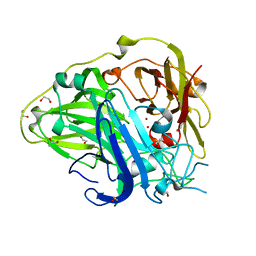 | | Crystal Structure of the reconstituted CotA | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, HYDROXIDE ION, ... | | Authors: | Bento, I, Silva, C.S, Chen, Z, Martins, L.O, Lindley, P.F, Soares, C.M. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanisms Underlying Dioxygen Reduction in Laccases. Structural and Modelling Studies Focusing on Proton Transfer.
Bmc Struct.Biol., 10, 2010
|
|
1ZE9
 
 | | Zinc-binding domain of Alzheimer's disease amyloid beta-peptide complexed with a zinc (II) cation | | Descriptor: | 16-mer from Alzheimer's disease amyloid Protein, ZINC ION | | Authors: | Zirah, S, Kozin, S.A, Mazur, A.K, Blond, A, Cheminant, M, Segalas-Milazzo, I, Debey, P, Rebuffat, S. | | Deposit date: | 2005-04-18 | | Release date: | 2005-05-03 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural changes of region 1-16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging.
J.Biol.Chem., 281, 2006
|
|
3VRT
 
 | | VDR ligand binding domain in complex with 2-Mehylidene-19,25,26,27-tetranor-1alpha,24-dihydroxyvitaminD3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R)-5-hydroxypentan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yoshimoto, N, Inaba, Y, Itoh, T, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
5H2R
 
 | | Crystal structure of T brucei phosphodiesterase B2 bound to compound 15b | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-(2-hydroxyethyloxy)-3-[2-(2-oxidanylideneimidazolidin-1-yl)ethoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, MAGNESIUM ION, Phosphodiesterase, ... | | Authors: | Noble, C.G. | | Deposit date: | 2016-10-17 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trypanosomal Phosphodiesterase B1 and B2 as a Potential Therapy for Human African Trypanosomiasis
To Be Published
|
|
6ZSE
 
 | | Human mitochondrial ribosome in complex with mRNA, A/P-tRNA and P/E-tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
5AXB
 
 | | Crystal structure of mouse SAHH complexed with noraristeromycin | | Descriptor: | (1S,2R,3S,4R)-4-(6-aminopurin-9-yl)cyclopentane-1,2,3-triol, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kusakabe, Y, Ishihara, M, Tanaka, N. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of mouse SAHH complexed with noraristeromycin
To Be Published
|
|
6ZSG
 
 | | Human mitochondrial ribosome in complex with mRNA, A-site tRNA, P-site tRNA and E-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
7UDH
 
 | | Integrin alpha IIB beta3 complex with BMS4-3 | | Descriptor: | (4-{[(5S)-3-(piperidin-4-yl)-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99998844 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
6TMC
 
 | | VIM-2_1dh-Triazole inhibitors with promising inhibitor effects against antibiotic resistance metallo-beta-lactamases | | Descriptor: | 4-[2-(phenylsulfonyl)ethyl]-5-(propan-2-yloxymethyl)-1~{H}-1,2,3-triazole, Beta-lactamase class B VIM-2, HYDROXIDE ION, ... | | Authors: | Leiros, H.-K.S, Christopeit, T. | | Deposit date: | 2019-12-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies of triazole inhibitors with promising inhibitor effects against antibiotic resistance metallo-beta-lactamases.
Bioorg.Med.Chem., 28, 2020
|
|
5QAF
 
 | | OXA-48 IN COMPLEX WITH COMPOUND 8c | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-fluorophenyl)benzoic acid, Beta-lactamase, ... | | Authors: | Lund, B.A, Leiros, H.K.S. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A focused fragment library targeting the antibiotic resistance enzyme - Oxacillinase-48: Synthesis, structural evaluation and inhibitor design.
Eur J Med Chem, 145, 2018
|
|
7UFH
 
 | | Integrin alpha IIB beta3 complex with fradafiban (Mn/Ca) | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-22 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.99768162 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
8BES
 
 | |
1L68
 
 | |
5GJK
 
 | | Crystal Structure of BAF47 and BAF155 Complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, SWI/SNF complex subunit SMARCC1, ... | | Authors: | Yan, L, Qian, C. | | Deposit date: | 2016-06-30 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Structural Insights into BAF47 and BAF155 Complex Formation.
J. Mol. Biol., 429, 2017
|
|
1ZRZ
 
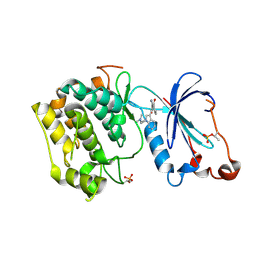 | | Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, Protein kinase C, iota | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M, Baedeker, M, Benda, C, Jestel, A, Brandstetter, H, Neuefeind, T, Blaesse, M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Atypical Protein Kinase C-iota Reveals Interaction Mode of Phosphorylation Site in Turn Motif
J.Mol.Biol., 352, 2005
|
|
7UCY
 
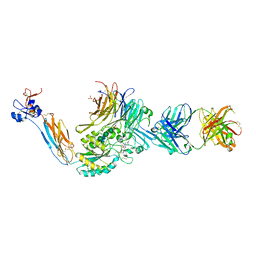 | | Integrin alpha IIB beta3 complex with gantofiban | | Descriptor: | (4-{[(5R)-3-(4-carbamimidoylphenyl)-2-oxo-1,3-oxazolidin-5-yl]methyl}piperazin-1-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.34996319 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
3IAE
 
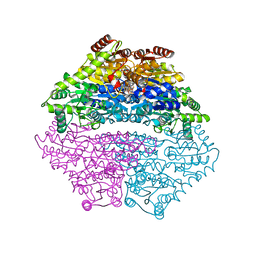 | | Structure of benzaldehyde lyase A28S mutant with benzoylphosphonate | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzaldehyde lyase, CALCIUM ION | | Authors: | Brandt, G.S, Petsko, G.A, Ringe, D, McLeish, M.J. | | Deposit date: | 2009-07-13 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-site engineering of benzaldehyde lyase shows that a point mutation can confer both new reactivity and susceptibility to mechanism-based inhibition.
J.Am.Chem.Soc., 132, 2010
|
|
2X40
 
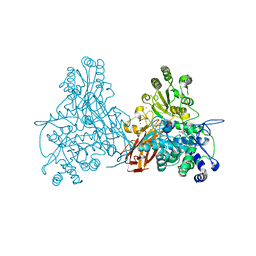 | |
9CE4
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 indazole inhibitor compound 6 | | Descriptor: | (1S,4r)-4-[(1P)-5-chloro-1-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-6-yl]-1-(oxetan-3-yl)piperidin-1-ium, 1,2-ETHANEDIOL, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L, Zebisch, M, Henry, C, Barker, J.J. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Discovery and Optimization of N-Heteroaryl Indazole LRRK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
4CUP
 
 | | Crystal structure of human BAZ2B in complex with fragment-1 N09421 | | Descriptor: | 4-Fluorobenzamidoxime, BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2B, METHANOL | | Authors: | Bradley, A.R, Liu, Y, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, von Delft, F. | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Human Baz2B in Complex with Fragment-1 N09421
To be Published
|
|
7UDG
 
 | | Integrin alpha IIB beta3 complex with lotrafiban | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.800069 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
6NHQ
 
 | |
