5U4Z
 
 | | Crystal structure of citrus MAF1 in space group P 31 2 1 | | Descriptor: | Repressor of RNA polymerase III transcription, SULFATE ION | | Authors: | Soprano, A.S, Giuseppe, P.O, Nascimento, A.F.Z, Benedetti, C.E, Murakami, M.T. | | Deposit date: | 2016-12-06 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of citrus MAF1 in space group P 31 2 1
To Be Published
|
|
4ASX
 
 | | Crystal structure of Activin receptor type-IIA (ACVR2A) kinase domain in complex with dihydro-Bauerine C | | Descriptor: | 1,2-ETHANEDIOL, 7,8-bis(chloranyl)-9-methyl-3,4-dihydro-2H-pyrido[3,4-b]indol-1-one, ACTIVIN RECEPTOR TYPE-2A | | Authors: | Williams, E, Chaikuad, A, Canning, P, Kochan, G, Mahajan, P, Cooper, C.D.O, Beltrami, A, Krojer, T, Pohl, B, Bracher, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A. | | Deposit date: | 2012-05-03 | | Release date: | 2012-05-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Activin Receptor Type-Iia (Acvr2A) Kinase Domain in Complex with a Beta- Carboline Inhibitor
To be Published
|
|
5OWC
 
 | | Indole-2 carboxamides as selective secreted phospholipase A2 type X (sPLA2-X) inhibitors | | Descriptor: | 3-[3-[2-aminocarbonyl-6-(trifluoromethyloxy)indol-1-yl]phenyl]propanoic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J.S, Roth, R.G, Knerr, L, Bodin, C, Pettersen, D. | | Deposit date: | 2017-08-31 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of a Series of Indole-2 Carboxamides as Selective Secreted Phospholipase A2Type X (sPLA2-X) Inhibitors.
Acs Med.Chem.Lett., 9, 2018
|
|
5UIJ
 
 | | X-ray structure of The FdtF N-formyltransferase from Salmonella enteric O60 in complex with TDP | | Descriptor: | 1,2-ETHANEDIOL, Formyltransferase, SODIUM ION, ... | | Authors: | Thoden, J.B, Woodford, C.R, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
5Z0Y
 
 | |
6DIC
 
 | | D276G DNA polymerase beta substrate complex with templating cytosine and incoming Fapy-dGTP analog | | Descriptor: | 1-[2-amino-5-(formylamino)-6-oxo-1,6-dihydropyrimidin-4-yl]-2,5-anhydro-1,3-dideoxy-6-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-D-ribo-hexitol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Freudenthal, B.D, Smith, M.R, Wilson, S.H, Beard, W.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | A guardian residue hinders insertion of a Fapy•dGTP analog by modulating the open-closed DNA polymerase transition.
Nucleic Acids Res., 47, 2019
|
|
4GYI
 
 | | Crystal structure of the Rio2 kinase-ADP/Mg2+-phosphoaspartate complex from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ferreira-Cerca, S, Sagar, V, Schafer, T, Diop, M, Wesseling, A.M, Lu, H, Chai, E, Hurt, E, LaRonde-LeBlanc, N. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7XEK
 
 | | SufS with D-cysteine for 30 min | | Descriptor: | (2~{S},4~{S})-2-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Visualizing thiazolidine ring formation in the reaction of D-cysteine and pyridoxal-5'-phosphate within L-cysteine desulfurase SufS.
Biochem.Biophys.Res.Commun., 754, 2025
|
|
3V9Y
 
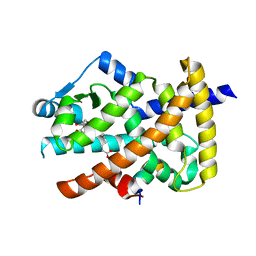 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | 4-{4-[({[(9aS)-8-acetyl-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-yl]carbonyl}amino)methyl]naphthalen-2-yl}butanoic acid, Peptide from Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substituents at the naphthalene C3 position of (-)-Cercosporamide derivatives significantly affect the maximal efficacy as PPAR(gamma) partial agonists
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5CT2
 
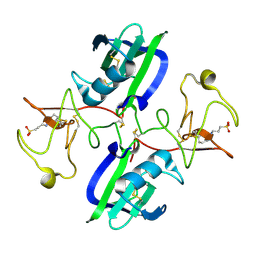 | | The structure of the NK1 fragment of HGF/SF complexed with CAPS | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5DHF
 
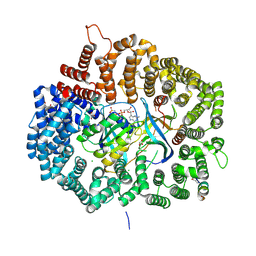 | |
3BJD
 
 | | Crystal structure of putative 3-oxoacyl-(acyl-carrier-protein) synthase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Putative 3-oxoacyl-(acyl-carrier-protein) synthase | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-03 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of putative 3-oxoacyl-(acyl-carrier-protein) synthase from Pseudomonas aeruginosa.
To be Published
|
|
7I1Y
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with 3632-JP-070-004 | | Descriptor: | (1s,3s)-3-amino-N-[4-(hydroxymethyl)-2-methylquinolin-8-yl]cyclobutane-1-carboxamide, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Marples, P.G, Godoy, A.S, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Fairhead, M, Lithgo, R.M, Balaji, G, Phelps, J, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Williams, E.P, Chandran, A.V, Walsh, M.A, Fearon, D, von Delft, F. | | Deposit date: | 2025-02-20 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3VD7
 
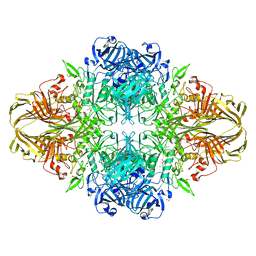 | | E. coli (lacZ) beta-galactosidase (N460S) in complex with galactotetrazole | | Descriptor: | (5R, 6S, 7S, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
7I1W
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with 3632-JP-070-010 | | Descriptor: | (1r,4r)-4-amino-N-[4-(hydroxymethyl)-2-methylquinolin-8-yl]cyclohexane-1-carboxamide, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Marples, P.G, Godoy, A.S, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Fairhead, M, Lithgo, R.M, Balaji, G, Phelps, J, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Williams, E.P, Chandran, A.V, Walsh, M.A, Fearon, D, von Delft, F. | | Deposit date: | 2025-02-20 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1YZ3
 
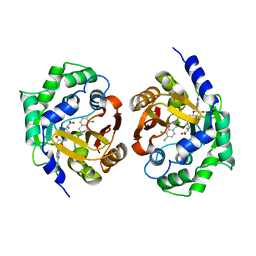 | | Structure of human pnmt complexed with cofactor product adohcy and inhibitor SK&F 64139 | | Descriptor: | 7,8-DICHLORO-1,2,3,4-TETRAHYDROISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wu, Q, Gee, C.L, Lin, F, Martin, J.L, Grunewald, G.L, McLeish, M.J. | | Deposit date: | 2005-02-27 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, mutagenic, and kinetic analysis of the binding of substrates and inhibitors of human phenylethanolamine N-methyltransferase
J.Med.Chem., 48, 2005
|
|
8CXJ
 
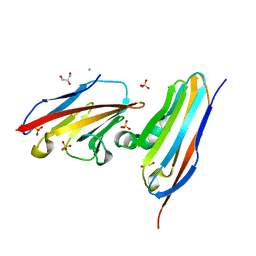 | |
1O0M
 
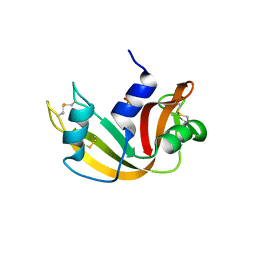 | | Ribonuclease A in complex with uridine-2'-phosphate | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Chrysina, E.D, Kosmopoulou, M.N, Vlassi, M. | | Deposit date: | 2003-02-24 | | Release date: | 2003-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of ribonuclease A complexed with adenylic and uridylic nucleotide inhibitors. Implications for structure-based design of ribonucleolytic inhibitors
PROTEIN SCI., 12, 2003
|
|
5OMM
 
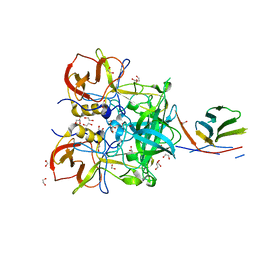 | |
6HTP
 
 | |
5J68
 
 | | Structure of Astrotactin-2, a conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Astrotactin-2, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Ni, T, Harlos, K, Gilbert, R.J.C. | | Deposit date: | 2016-04-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (5.221 Å) | | Cite: | Structure of astrotactin-2: a conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development.
Open Biology, 6, 2016
|
|
6OJP
 
 | | Structure of glycolipid alpha-GSA[8,6P] in complex with mouse CD1d | | Descriptor: | (5R,6S,7S)-5,6-dihydroxy-7-(octanoylamino)-N-(6-phenylhexyl)-8-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}octanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6HW6
 
 | | Yeast 20S proteasome in complex with 20 | | Descriptor: | MAGNESIUM ION, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
4AA9
 
 | | Camel chymosin at 1.6A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHYMOSIN, GLYCEROL, ... | | Authors: | Langholm Jensen, J, Molgaard, A, Navarro Poulsen, J.C, van den Brink, J.M, Harboe, M, Simonsen, J.B, Qvist, K.B, Larsen, S. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Camel and Bovine Chymosin: The Relationship between Their Structures and Cheese-Making Properties.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4DHY
 
 | | Crystal structure of human glucokinase in complex with glucose and activator | | Descriptor: | Glucokinase, N,N-dimethyl-5-({2-methyl-6-[(5-methylpyrazin-2-yl)carbamoyl]-1-benzofuran-4-yl}oxy)pyrimidine-2-carboxamide, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
