7HFQ
 
 | |
7CMF
 
 | | Crystal structure of human P-cadherin REC12 (monomer) in complex with 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine (inhibitor) | | Descriptor: | 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3 | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulation of cadherin dimerization by chemical fragments as a trigger to inhibit cell adhesion
Commun Biol, 4, 2021
|
|
7HFW
 
 | |
7HE6
 
 | |
6CGZ
 
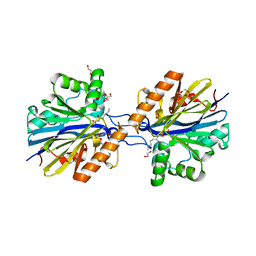 | | Structure of the Quorum Quenching lactonase from Alicyclobacillus acidoterrestris bound to C6-AHL | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, COBALT (II) ION, ... | | Authors: | Bergonzi, C, Schwab, M, Naik, T, Daude, D, Chabriere, E, Elias, M. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties.
Sci Rep, 8, 2018
|
|
7HEU
 
 | |
7HFT
 
 | |
3U6B
 
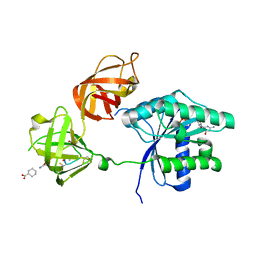 | | Ef-tu (escherichia coli) in complex with nvp-ldi028 | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Palestrant, D.J. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Antibacterial optimization of 4-aminothiazolyl analogues of the natural product GE2270 A: identification of the cycloalkylcarboxylic acids.
J.Med.Chem., 54, 2011
|
|
6CIG
 
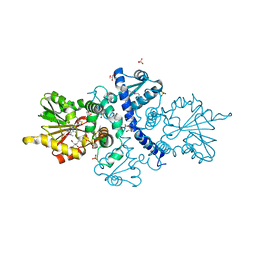 | | CRYSTAL STRUCTURE ANALYSIS OF SELENOMETHIONINE SUBSTITUTED ISOFLAVONE O-METHYLTRANSFERASE | | Descriptor: | GLYCEROL, Isoflavone-7-O-methyltransferase 8, N-(TRIS(HYDROXYMETHYL)METHYL)-3-AMINOPROPANESULFONIC ACID, ... | | Authors: | Zubieta, C, Dixon, R.A, Shabalin, I.G, Kowiel, M, Porebski, P.J, Noel, J.P. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat. Struct. Biol., 8, 2001
|
|
3MNR
 
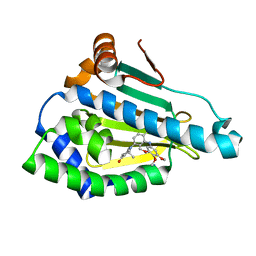 | | Crystal Structure of Benzamide SNX-1321 bound to Hsp90 | | Descriptor: | 2-[(3,4,5-trimethoxyphenyl)amino]-4-(2,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)benzamide, Heat shock protein HSP 90-alpha | | Authors: | Veal, J.M, Fadden, P, Huang, K.H, Rice, J, Hall, S.E, Haytstead, T.A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Chemoproteomics to Drug Discovery: Identification of a Clinical Candidate Targeting Hsp90.
Chem.Biol., 17, 2010
|
|
6QFV
 
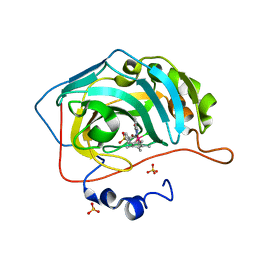 | | Human carbonic anhydrase II with bound IrCp* complex (cofactor 8) to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[2-(9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1(6),2,4-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Chemical Optimization of Whole-Cell Transfer Hydrogenation Using Carbonic Anhydrase as Host Protein.
Acs Catalysis, 9, 2019
|
|
7HEJ
 
 | |
7HDP
 
 | |
7HEZ
 
 | |
7HDA
 
 | |
5SXL
 
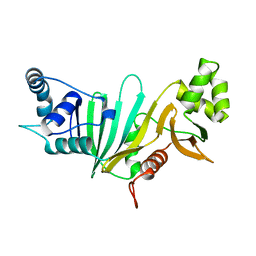 | |
7MM2
 
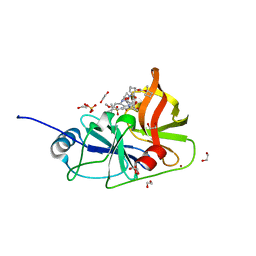 | | Crystal structure of HCV NS3/4A protease in complex with NR02-61 | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4a protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7HEC
 
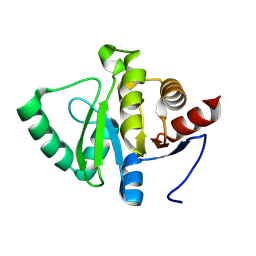 | |
6TIA
 
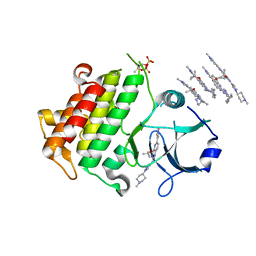 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 4-(1-methylcyclopropyl)oxy-~{N}-[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]-6-(1-methylpyrazol-4-yl)pyrido[3,2-d]pyrimidin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Degorce, S.L. | | Deposit date: | 2019-11-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Improving metabolic stability and removing aldehyde oxidase liability in a 5-azaquinazoline series of IRAK4 inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
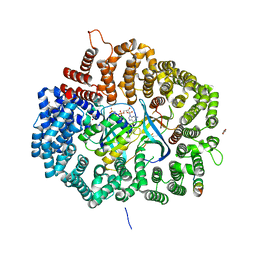
5DH9
 
 | |
7HDF
 
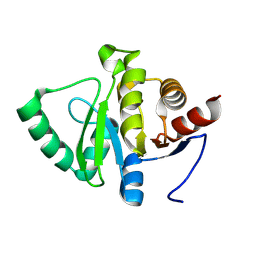 | |
8SG8
 
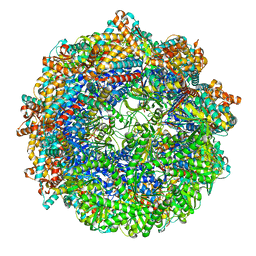 | | CCT G beta 5 complex closed state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-10-25 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
7MMG
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-58 | | Descriptor: | 1-(trifluoromethyl)cyclobutyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, ARGININE, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
6TMB
 
 | |
7HDV
 
 | |
