3UNT
 
 | | tRNA-guanine transglycosylase E339Q mutant | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Jakobi, S, Heine, A, Klebe, G. | | Deposit date: | 2011-11-16 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Hot-spot analysis to dissect the functional protein-protein interface of a tRNA-modifying enzyme.
Proteins, 82, 2014
|
|
1PQV
 
 | | RNA polymerase II-TFIIS complex | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | Authors: | Kettenberger, H, Armache, K.-J, Cramer, P. | | Deposit date: | 2003-06-19 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Architecture of the RNA Polymerase II-TFIIS Complex and Implications for mRNA Cleavage
Cell(Cambridge,Mass.), 114, 2003
|
|
5M0H
 
 | |
1NT9
 
 | | Complete 12-subunit RNA polymerase II | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6 KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 19 KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II LARGEST SUBUNIT, ... | | Authors: | Armache, K.-J, Kettenberger, H, Cramer, P. | | Deposit date: | 2003-01-29 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Architecture of initiation-competent 12-subunit RNA polymerase II
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1D6T
 
 | | RNASE P PROTEIN FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | RIBONUCLEASE P | | Authors: | Spitzfaden, C. | | Deposit date: | 1999-10-15 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of ribonuclease P protein from Staphylococcus aureus reveals a unique binding site for single-stranded RNA.
J.Mol.Biol., 295, 2000
|
|
7AFO
 
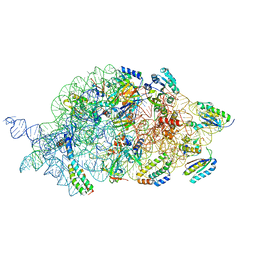 | | Bacterial 30S ribosomal subunit assembly complex state B (body domain) | | Descriptor: | 16SrRNA (body domain of the 30S ribosome), 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7ZPJ
 
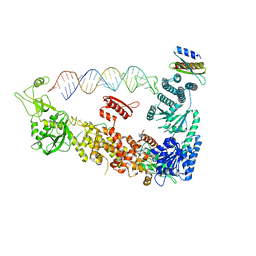 | | Mammalian Dicer in the "pre-dicing state" with pre-miR-15a substrate and TARBP2 subunit | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 isoform 1 | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-04-27 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
4XWT
 
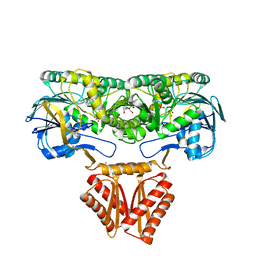 | | Crystal structure of RNase J complexed with UMP | | Descriptor: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | Deposit date: | 2015-01-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
7ZPK
 
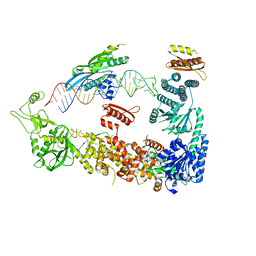 | | Mammalian Dicer in the "pre-dicing state" with pre-miR-15a substrate and TARBP2 subunit | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
6HLS
 
 | | Yeast apo RNA polymerase I* | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
5XOG
 
 | | RNA Polymerase II elongation complex bound with Spt5 KOW5 and Elf1 | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA (30-MER), DNA (39-MER), ... | | Authors: | Ehara, H, Shirouzu, M, Sekine, S. | | Deposit date: | 2017-05-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the complete elongation complex of RNA polymerase II with basal factors
Science, 357, 2017
|
|
3M7N
 
 | |
3M85
 
 | |
7ZPI
 
 | | Mammalian Dicer in the "dicing state" with pre-miR-15a substrate | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (5.91 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
5OT2
 
 | | RNA polymerase II elongation complex in the presence of 3d-Napht-A | | Descriptor: | 2-ethyl-7-methoxy-naphthalene, DNA non-template strand, DNA template strand, ... | | Authors: | Malvezzi, S, Farnung, L, Aloisi, C, Angelov, T, Cramer, P, Sturla, S.J. | | Deposit date: | 2017-08-19 | | Release date: | 2017-11-01 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of RNA polymerase II stalling by DNA alkylation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7NVT
 
 | | RNA polymerase II core pre-initiation complex with closed promoter DNA in distal position | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7NVU
 
 | | RNA polymerase II core pre-initiation complex with open promoter DNA | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7NVS
 
 | | RNA polymerase II core pre-initiation complex with closed promoter DNA in proximal position | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
6J6G
 
 | | Cryo-EM structure of the yeast B*-a2 complex at an average resolution of 3.2 angstrom | | Descriptor: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6J6H
 
 | | Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom | | Descriptor: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6J6N
 
 | | Cryo-EM structure of the yeast B*-b1 complex at an average resolution of 3.86 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6J6Q
 
 | | Cryo-EM structure of the yeast B*-b2 complex at an average resolution of 3.7 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6L8U
 
 | | Crystal structure of human BCDIN3D in complex with SAH | | Descriptor: | RNA 5'-monophosphate methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, Y, Martinez, A, Yamashita, S, Tomita, K. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.925 Å) | | Cite: | Crystal structure of human cytoplasmic tRNAHis-specific 5'-monomethylphosphate capping enzyme.
Nucleic Acids Res., 48, 2020
|
|
7AFL
 
 | | Bacterial 30S ribosomal subunit assembly complex state D (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16SrRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
5GAO
 
 | | Head region of the yeast spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | Pre-mRNA-splicing factor 8, Pre-mRNA-splicing helicase BRR2, Saccharomyces cerevisiae strain UOA_M2 chromosome 5 sequence, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
