4BWU
 
 | | Three-dimensional structure of the K109A mutant of Paracoccus pantotrophus pseudoazurin at pH 5.5 | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN, SULFATE ION | | Authors: | Freire, F, Mestre, A, Pinho, J, Najmudin, S, Bonifacio, C, Pauleta, S.R, Romao, M.J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Exploring the Surface Determinants of Paracoccus Pantotrophus Pseudoazurin
To be Published
|
|
5GNY
 
 | | The structure of WT Bgl6 | | Descriptor: | Beta-glucosidase, beta-D-glucopyranose | | Authors: | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | Deposit date: | 2016-07-25 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
5UCR
 
 | |
4CTG
 
 | | The limits of structural plasticity in a picornavirus capsid revealed by a massively expanded equine rhinitis A virus particle | | Descriptor: | P1 | | Authors: | Bakker, S.E, Groppelli, E, Pearson, A.R, Stockley, P.G, Rowlands, D.J, Ranson, N.A. | | Deposit date: | 2014-03-13 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Limits of Structural Plasticity in a Picornavirus Capsid Revealed by a Massively Expanded Equine Rhinitis a Virus Particle.
J.Virol., 88, 2014
|
|
4CTF
 
 | | The limits of structural plasticity in a picornavirus capsid revealed by a massively expanded equine rhinitis A virus particle | | Descriptor: | EQUINE RHINITIS A VIRUS, P1, VP1 | | Authors: | Bakker, S.E, Groppelli, E, Pearson, A.R, Stockley, P.G, Rowlands, D.J, Ranson, N.A. | | Deposit date: | 2014-03-13 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Limits of Structural Plasticity in a Picornavirus Capsid Revealed by a Massively Expanded Equine Rhinitis a Virus Particle.
J.Virol., 88, 2014
|
|
5H0K
 
 | |
5H0J
 
 | |
4FOW
 
 | |
4BWT
 
 | | Three-dimensional structure of Paracoccus pantotrophus pseudoazurin at pH 6.5 | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN, SULFATE ION | | Authors: | Freire, F, Mestre, A, Pinho, J, Najmudin, S, Bonifacio, C, Pauleta, S.R, Romao, M.J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Exploring the Surface Determinants of Paracoccus Pantotrophus Pseudoazurin
To be Published
|
|
4BXV
 
 | | Three-dimensional structure of the mutant K109A of Paracoccus pantotrophus pseudoazurin at pH 7.0 | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN, SULFATE ION | | Authors: | Freire, F, Mestre, A, Pinho, J, Najmudin, S, Bonifacio, C, Pauleta, S.R, Romao, M.J. | | Deposit date: | 2013-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Exploring the Surface Determinants of Paracoccus Pantotrophus Pseudoazurin
To be Published
|
|
4DOQ
 
 | | Crystal structure of the complex of Porcine Pancreatic Trypsin with 1/2SLPI | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Antileukoproteinase, CALCIUM ION, ... | | Authors: | Fukushima, K, Takimoto-Kamimura, M. | | Deposit date: | 2012-02-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure basis 1/2SLPI and porcine pancreas trypsin interaction
J.SYNCHROTRON RADIAT., 20, 2013
|
|
5UTR
 
 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-butyryl-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]butanamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2017-02-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational flexibility of the glycosidase NagZ allows it to bind structurally diverse inhibitors to suppress beta-lactam antibiotic resistance.
Protein Sci., 26, 2017
|
|
4B7M
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
5TS2
 
 | |
7EST
 
 | |
5UL6
 
 | | The molecular mechanisms by which NS1 of the 1918 Spanish influenza A virus hijack host protein-protein interactions | | Descriptor: | Adapter molecule crk, Proline-rich motif of nonstructural protein 1 of influenza a virus | | Authors: | Shen, Q, Zeng, D, Zhao, B, Li, P, Cho, J.H. | | Deposit date: | 2017-01-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Molecular Mechanisms Underlying the Hijack of Host Proteins by the 1918 Spanish Influenza Virus.
ACS Chem. Biol., 12, 2017
|
|
5VH7
 
 | | Structure and dynamics of RNA repeat expansions that cause Huntington's Disease and myotonic dystrophy type 1 | | Descriptor: | RNA (5'-R(*GP*AP*CP*CP*AP*GP*CP*AP*G)-3') | | Authors: | Chen, J.L, VanEtten, D.M, Fountain, M.A, Yildirim, I, Disney, M.D. | | Deposit date: | 2017-04-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of RNA Repeat Expansions That Cause Huntington's Disease and Myotonic Dystrophy Type 1.
Biochemistry, 56, 2017
|
|
5VH8
 
 | | Structure and dynamics of RNA repeat expansions that cause Huntington's Disease and myotonic dystrophy type 1 | | Descriptor: | RNA (5'-R(*GP*AP*CP*CP*UP*GP*CP*UP*GP*CP*UP*GP*GP*UP*C)-3') | | Authors: | Chen, J.L, VanEtten, D.M, Fountain, M.A, Yildirim, I, Disney, M.D. | | Deposit date: | 2017-04-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of RNA Repeat Expansions That Cause Huntington's Disease and Myotonic Dystrophy Type 1.
Biochemistry, 56, 2017
|
|
7F30
 
 | | Crystal structure of OxdB E85A in complex with Z-2- (3-bromophenyl) propanal oxime | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase, Z-2-(3-bromophenyl) propanal oxime | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
7E4W
 
 | |
7JLN
 
 | |
7JLK
 
 | |
5PTI
 
 | |
5OPT
 
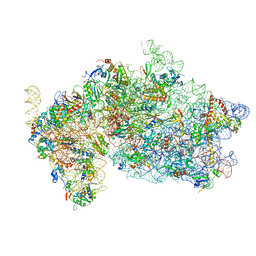 | | Structure of KSRP in context of Trypanosoma cruzi 40S | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, putative, ... | | Authors: | Brito Querido, J, Mancera-Martinez, E, Vicens, Q, Bochler, A, Chicher, J, Simonetti, A, Hashem, Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein.
Structure, 25, 2017
|
|
5NWN
 
 | | Deinococcus radiodurans BphP PAS-GAF-PHY Y263F mutant, dark | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Takala, H, Westehoff, S, Ihalainen, J.A. | | Deposit date: | 2017-05-07 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | On the (un)coupling of the chromophore, tongue interactions, and overall conformation in a bacterial phytochrome.
J. Biol. Chem., 293, 2018
|
|
