2RBY
 
 | | 1-methyl-5-imidazolecarboxaldehyde in complex with Cytochrome C Peroxidase W191G | | Descriptor: | 1-methyl-1H-imidazole-5-carbaldehyde, Cytochrome C Peroxidase, PHOSPHATE ION, ... | | Authors: | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
9BNL
 
 | | Cryo-EM structure of rhesus antibody 6070-a.01 in complex with HIV-1 Env trimer Q23.17 MD39 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6070-a.01 heavy chain, ... | | Authors: | Roark, R.S, Shapiro, L, Kwong, P.D. | | Deposit date: | 2024-05-02 | | Release date: | 2025-05-07 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and genetic basis of HIV-1 envelope V2 apex recognition by rhesus broadly neutralizing antibodies.
J.Exp.Med., 222, 2025
|
|
9BNM
 
 | | Cryo-EM structure of rhesus antibody 44715-a.01 in complex with HIV-1 Env BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 44715-a.01 light chain, ... | | Authors: | Roark, R.S, Shapiro, L, Kwong, P.D. | | Deposit date: | 2024-05-02 | | Release date: | 2025-05-07 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural and genetic basis of HIV-1 envelope V2 apex recognition by rhesus broadly neutralizing antibodies.
J.Exp.Med., 222, 2025
|
|
9BNP
 
 | | Cryo-EM structure of rhesus antibody V033-a.01 in complex with HIV-1 Env BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein Gp120, ... | | Authors: | Roark, R.S, Shapiro, L, Kwong, P.D. | | Deposit date: | 2024-05-02 | | Release date: | 2025-05-07 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural and genetic basis of HIV-1 envelope V2 apex recognition by rhesus broadly neutralizing antibodies.
J.Exp.Med., 222, 2025
|
|
3KST
 
 | |
6A35
 
 | |
5HU1
 
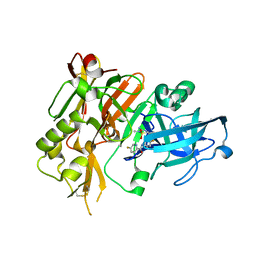 | | BACE1 in complex with (R)-N-(3-(3-amino-2,5-dimethyl-1,1-dioxido-5,6-dihydro-2H-1,2,4-thiadiazin-5-yl)-4-fluorophenyl)-5-fluoropicolinamide | | Descriptor: | Beta-secretase 1, N-{3-[(5R)-3-amino-2,5-dimethyl-1,1-dioxido-5,6-dihydro-2H-1,2,4-thiadiazin-5-yl]-4-fluorophenyl}-5-fluoropyridine-2-carboxamide | | Authors: | Orth, P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)-A beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer's Disease.
J. Med. Chem., 59, 2016
|
|
3CGG
 
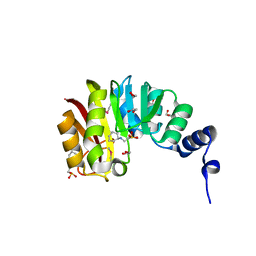 | |
9BNK
 
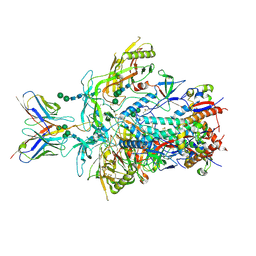 | | Cryo-EM structure of rhesus antibody V031-a.01 in complex with HIV-1 Env BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein Gp41, ... | | Authors: | Roark, R.S, Shapiro, L, Kwong, P.D. | | Deposit date: | 2024-05-02 | | Release date: | 2025-05-07 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and genetic basis of HIV-1 envelope V2 apex recognition by rhesus broadly neutralizing antibodies.
J.Exp.Med., 222, 2025
|
|
3CVQ
 
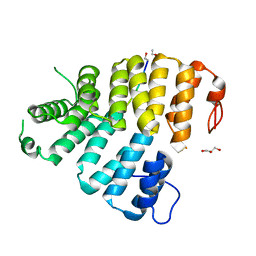 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (7-SKL) | | Descriptor: | GLYCEROL, PTS1 peptide 7-SKL (Ac-SNRWSKL), Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
5DHQ
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a novel inhibitor | | Descriptor: | 8-[(2-{[2-(3-bromophenyl)ethyl]amino}-2-oxoethyl)sulfanyl]adenosine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Paoletti, J, Assairi, L, Huteau, V, Pochet, S, Labesse, G. | | Deposit date: | 2015-08-31 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | 8-Thioalkyl-adenosine derivatives inhibit Listeria monocytogenes NAD kinase through a novel binding mode.
Eur.J.Med.Chem., 124, 2016
|
|
4ZPG
 
 | |
9BTH
 
 | | Rhesus Fab 42056-a.01 in complex with CAP256SU.wk34 RnS SOSIP Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-05-15 | | Release date: | 2025-06-11 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and genetic basis of HIV-1 envelope V2 apex recognition by rhesus broadly neutralizing antibodies.
J.Exp.Med., 222, 2025
|
|
5U1C
 
 | |
9BTV
 
 | | Cryo-EM structure of rhesus antibody T646-a.01 in complex with HIV-1 Env trimer Q23.17 MD39 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Roark, R.S, Shapiro, L, Kwong, P.D. | | Deposit date: | 2024-05-15 | | Release date: | 2025-07-02 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and genetic basis of HIV-1 envelope V2 apex recognition by rhesus broadly neutralizing antibodies.
J.Exp.Med., 222, 2025
|
|
6F6R
 
 | | Crystal structure of human Caspase-1 with N-{3-[1-((S)-2-Hydroxy-5-oxo-tetrahydro-furan-3-ylcarbamoyl)-ethyl]-1-methyl-2,4-dioxo-1,2,3,4-tetrahydro-pyrimidin-5-yl}-4-(quinoxalin-2-ylamino)-benzamide | | Descriptor: | (3~{S})-3-[[(2~{R})-2-[3-methyl-2,6-bis(oxidanylidene)-5-[[4-(quinoxalin-2-ylamino)phenyl]carbonylamino]pyrimidin-1-yl]propanoyl]amino]-4-oxidanyl-butanoic acid, Caspase-1, SULFATE ION | | Authors: | Fournier, J.F, Clary, L, Chambon, S, Dumais, L, Harris, C.S, Millois-Barbuis, C, Pierre, R, Talano, S, Thoreau, E, Aubert, J, Aurelly, M, Bouix-Peter, C, Brethon, A, Chantalat, L, Christin, O, Comino, C, El-Bazbouz, G, Ghilini, A.L, Isabet, T, Lardy, C, Luzy, A.P, Mathieu, C, Mebrouk, K, Orfila, D, Pascau, J, Reverse, K, Roche, D, Rodeschini, V, Hennequin, L.F. | | Deposit date: | 2017-12-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Drug Design of Topically Administered Caspase 1 Inhibitors for the Treatment of Inflammatory Acne.
J. Med. Chem., 61, 2018
|
|
6TTC
 
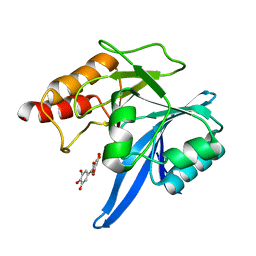 | | Haddock model of NDM-1/myricetin complex | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
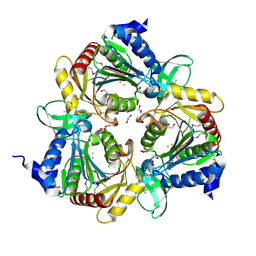
3CPX
 
 | |
7XM9
 
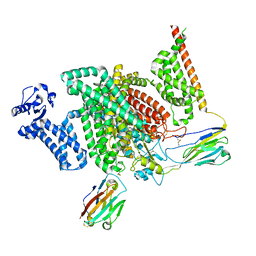 | | Cryo-EM structure of human NaV1.7/beta1/beta2-XEN907 | | Descriptor: | (7~{R})-1'-pentylspiro[6~{H}-furo[3,2-f][1,3]benzodioxole-7,3'-indole]-2'-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | zhang, J.T, Jiang, D.H. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4LGN
 
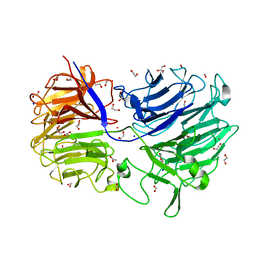 | | The structure of Acidothermus cellulolyticus family 74 glycoside hydrolase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cellulose-binding, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2013-06-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Acidothermus cellulolyticus family 74 glycoside hydrolase at 1.82 angstrom resolution.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3LIX
 
 | | crystal structure of htlv protease complexed with the inhibitor KNI-10729 | | Descriptor: | N-{(1S,2S)-1-benzyl-3-[(4R)-5,5-dimethyl-4-{[(1R)-1,2,2-trimethylpropyl]carbamoyl}-1,3-thiazolidin-3-yl]-2-hydroxy-3-oxopropyl}-3-methyl-N~2~-{(2S)-2-[(morpholin-4-ylacetyl)amino]-2-phenylacetyl}-L-valinamide, Protease, ZINC ION | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
6AXW
 
 | |
6L4P
 
 | | Crystal structure of the complex between the axonemal outer-arm dynein light chain-1 and microtubule binding domain of gamma heavy chain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dynein light chain 1, axonemal, ... | | Authors: | Toda, A, Nishikawa, Y, Tanaka, H, Yagi, T, Kurisu, G. | | Deposit date: | 2019-10-19 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | The complex of outer-arm dynein light chain-1 and the microtubule-binding domain of the gamma heavy chain shows how axonemal dynein tunes ciliary beating.
J.Biol.Chem., 295, 2020
|
|
9FNH
 
 | | The glycoside hydrolase family 71 (GH71) member AnGH71C from Aspergillus nidulans in complex with nigerotetraose. | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Glycoside hydrolase family 71, ... | | Authors: | Mazurkewich, S, Widen, T, Branden, G, Larsbrink, J. | | Deposit date: | 2024-06-10 | | Release date: | 2025-06-25 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and biochemical basis for activity of Aspergillus nidulans alpha-1,3-glucanases from glycoside hydrolase family 71.
Commun Biol, 8, 2025
|
|
1LOK
 
 | | The 1.20 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica Complexed with Tris: A Tale of Buffer Inhibition | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bacterial leucyl aminopeptidase, SODIUM ION, ... | | Authors: | Desmarais, W.T, Bienvenue, D.L, Bzymek, K.P, Holz, R.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.20 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica Complexed with Tris A tale of Buffer Inhibition
Structure, 10, 2002
|
|
