5SRL
 
 | |
5SRM
 
 | |
5SRP
 
 | |
5SRQ
 
 | |
5SRV
 
 | |
5SRU
 
 | |
5SRT
 
 | |
5E0Y
 
 | |
2QTM
 
 | |
4M4P
 
 | | Crystal structure of EPHA4 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-A receptor 4 | | Authors: | Xu, K, Tsvetkova-Robev, D, Xu, Y, Goldgur, Y, Chan, Y.-P, Himanen, J.P, Nikolov, D.B. | | Deposit date: | 2013-08-07 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Insights into Eph receptor tyrosine kinase activation from crystal structures of the EphA4 ectodomain and its complex with ephrin-A5.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4QRR
 
 | | Crystal Structure of HLA B*3501-IPS in complex with a Delta-Beta TCR, clone 12 TCR | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Chabrol, E, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
J.Exp.Med., 211, 2014
|
|
3LC9
 
 | |
206D
 
 | |
4NJS
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL008 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-carbamoylphenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
5DYF
 
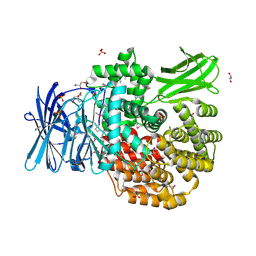 | | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid | | Descriptor: | Aminopeptidase N, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nocek, B, Joachimiak, A, Vassiliou, S, Berlicki, L, Mucha, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid
To Be Published
|
|
1QE5
 
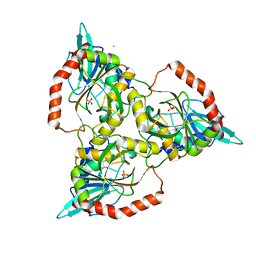 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM CELLULOMONAS SP. IN COMPLEX WITH PHOSPHATE | | Descriptor: | CALCIUM ION, PENTOSYLTRANSFERASE, PHOSPHATE ION | | Authors: | Tebbe, J, Bzowska, A, Wielgus-Kutrowska, B, Schroeder, W, Kazimierczuk, Z, Shugar, D, Saenger, W, Koellner, G. | | Deposit date: | 1999-07-13 | | Release date: | 1999-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the purine nucleoside phosphorylase (PNP) from Cellulomonas sp. and its implication for the mechanism of trimeric PNPs.
J.Mol.Biol., 294, 1999
|
|
4JGH
 
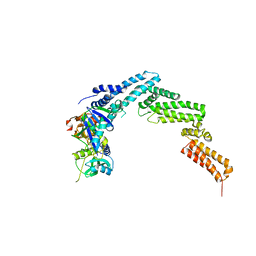 | | Structure of the SOCS2-Elongin BC complex bound to an N-terminal fragment of Cullin5 | | Descriptor: | Cullin-5, Suppressor of cytokine signaling 2, Transcription elongation factor B polypeptide 1, ... | | Authors: | Kim, Y.K, Kwak, M.J, Ku, B, Suh, H.Y, Joo, K, Lee, J, Jung, J.U, Oh, B.H. | | Deposit date: | 2013-03-01 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of intersubunit recognition in elongin BC-cullin 5-SOCS box ubiquitin-protein ligase complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3FPX
 
 | | Native fungus laccase from Trametes hirsuta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Stepanova, E.V, Cherkashin, E.A, Kurzeev, S.A, Strokopytov, B.V, Lamzin, V.S, Koroleva, O.V. | | Deposit date: | 2009-01-06 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of native laccase from Coriolus hirsutus at 1.8 A resolution
To be Published
|
|
1HRT
 
 | |
3F7H
 
 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2008-11-09 | | Release date: | 2009-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Orally bioavailable antagonists of inhibitor of apoptosis proteins based on an azabicyclooctane scaffold
J.Med.Chem., 52, 2009
|
|
4J4P
 
 | | The complex of human IgE-Fc with two bound Fab fragments | | Descriptor: | Ig epsilon chain C region, Immunoglobulin G Fab Fragment Heavy Chain, Immunoglobulin G Fab Fragment Light Chain, ... | | Authors: | Drinkwater, N, Sutton, B.J. | | Deposit date: | 2013-02-07 | | Release date: | 2014-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Human immunoglobulin E flexes between acutely bent and extended conformations.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1IXO
 
 | | Enzyme-analogue substrate complex of Pyridoxine 5'-Phosphate Synthase | | Descriptor: | Pyridoxine 5'-Phosphate synthase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
1BB7
 
 | |
4F20
 
 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION, peptide from Dermokine | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
1VEB
 
 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 5 | | Descriptor: | (3R,4R)-N-{4-[4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-BENZYLOXY]-AZEPAN-3-YL}-ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.-G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
