8Y64
 
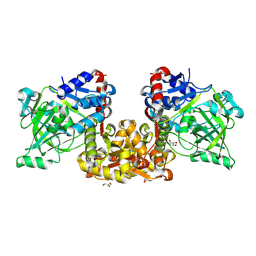 | | Crystal structure of open state ferulic acid decarboxylase from Saccharomyces cerevisiae, F397V/I398L/T438P/P441V mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ferulic acid decarboxylase 1, GLYCEROL | | Authors: | Feng, Y.B, Song, X, Zhu, X.N. | | Deposit date: | 2024-02-01 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Integrated strategies for engineering ferulic acid decarboxylase with tunnel conformation and substrate pocket for adapting non-natural substrates.
Biochem Eng J, 213, 2025
|
|
5IC1
 
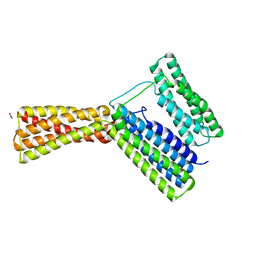 | | Structural analysis of a talin triple domain module, E1794Y, E1797Y, Q1801Y mutant | | Descriptor: | 1,2-ETHANEDIOL, Talin-1 | | Authors: | Wu, J, Chang, Y.-C.E, Zhang, H, Huang, Q.-Q. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration.
Structure, 24, 2016
|
|
3GH9
 
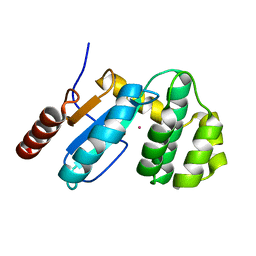 | | Crystal structure of EDTA-treated BdbD (Oxidised) | | Descriptor: | 1,2-ETHANEDIOL, Disulfide bond formation protein D, UNKNOWN ATOM OR ION | | Authors: | Crow, A, Lewin, A, Hederstedt, L, Le-Brun, N.E. | | Deposit date: | 2009-03-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structure and Biophysical Properties of Bacillus subtilis BdbD: AN OXIDIZING THIOL:DISULFIDE OXIDOREDUCTASE CONTAINING A NOVEL METAL SITE
J.Biol.Chem., 284, 2009
|
|
9OPB
 
 | | Herpes simplex virus type 1 (HSV-1) D-capsid pUL6 portal protein turrets, decamer | | Descriptor: | Capsid portal protein | | Authors: | Crofut, E.H, Kashyap, S, Stevens, A, Jih, J, Liu, Y.-T, Zhou, Z.H. | | Deposit date: | 2025-05-17 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a new capsid form and comparison with A-, B-, and C-capsids clarify herpesvirus assembly.
J.Virol., 99, 2025
|
|
9F63
 
 | |
6T3L
 
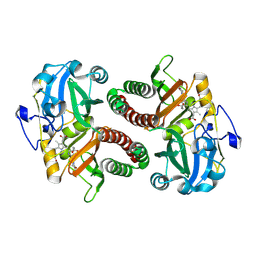 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome in dark state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
6T3U
 
 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome 1ps after photoexcitation | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
9OP5
 
 | | Herpes simplex virus type 1 (HSV-1) B-capsid pUL6 portal protein, dodecameric complex | | Descriptor: | Capsid portal protein, Capsid scaffolding protein | | Authors: | Crofut, E.H, Kashyap, S, Stevens, A, Jih, J, Liu, Y.-T, Zhou, Z.H. | | Deposit date: | 2025-05-16 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a new capsid form and comparison with A-, B-, and C-capsids clarify herpesvirus assembly.
J.Virol., 99, 2025
|
|
9OPC
 
 | | Herpes simplex virus type 1 (HSV-1) C-capsid pUL6 portal protein, dodecameric complex | | Descriptor: | Capsid portal protein | | Authors: | Crofut, E.H, Kashyap, S, Stevens, A, Jih, J, Liu, Y.-T, Zhou, Z.H. | | Deposit date: | 2025-05-18 | | Release date: | 2025-05-28 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of a new capsid form and comparison with A-, B-, and C-capsids clarify herpesvirus assembly.
J.Virol., 99, 2025
|
|
5BSV
 
 | |
6IL9
 
 | | One Glycerol complexed Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi | | Descriptor: | Fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi in complex with 1 glycerol, GLYCEROL, ZINC ION | | Authors: | Choi, M.Y, Kang, L.W, Ho, T.H, Nguyen, D.Q, Lee, I.H, Lee, J.H, Park, Y.S, Park, H.J. | | Deposit date: | 2018-10-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72005355 Å) | | Cite: | Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi
To Be Published
|
|
3GBE
 
 | | Crystal structure of the isomaltulose synthase SmuA from Protaminobacter rubrum in complex with the inhibitor deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, CITRATE ANION, ... | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants of product specificity of sucrose isomerases
Febs Lett., 583, 2009
|
|
9OP8
 
 | | Herpes simplex virus type 1 (HSV-1) D-capsid pUL6 portal protein, dodecameric complex | | Descriptor: | Capsid portal protein | | Authors: | Crofut, E.H, Kashyap, S, Stevens, A, Jih, J, Liu, Y.-T, Zhou, Z.H. | | Deposit date: | 2025-05-17 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a new capsid form and comparison with A-, B-, and C-capsids clarify herpesvirus assembly.
J.Virol., 99, 2025
|
|
5BSY
 
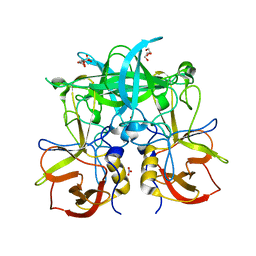 | |
6I5Z
 
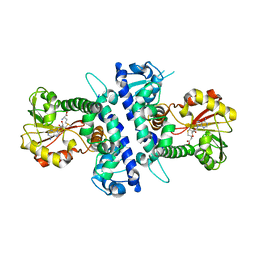 | | Papaver somniferum O-methyltransferase | | Descriptor: | O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE | | Authors: | Davies, G.J, Cabry, M.P, Offen, W.A. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
7X42
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP0, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
5ESB
 
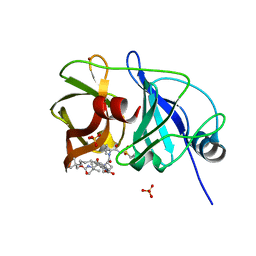 | | Crystal structure of a genotype 1a/3a chimeric HCV NS3/4A protease in complex with Vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Schiffer, C.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.
J.Am.Chem.Soc., 138, 2016
|
|
3EAM
 
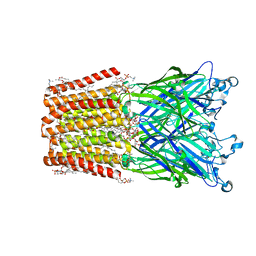 | | An open-pore structure of a bacterial pentameric ligand-gated ion channel | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Bocquet, N, Nury, H, Baaden, M, Le Poupon, C, Changeux, J.P, Delarue, M, Corringer, P.J. | | Deposit date: | 2008-08-26 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation.
Nature, 457, 2009
|
|
9OP4
 
 | | Herpes simplex virus type 1 (HSV-1) A-capsid pUL6 portal protein, dodecameric complex | | Descriptor: | Capsid portal protein, Capsid scaffolding protein | | Authors: | Crofut, E.H, Kashyap, S, Stevens, A, Jih, J, Liu, Y.-T, Zhou, Z.H. | | Deposit date: | 2025-05-16 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a new capsid form and comparison with A-, B-, and C-capsids clarify herpesvirus assembly.
J.Virol., 99, 2025
|
|
5HWJ
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase | | Descriptor: | FORMIC ACID, GLYCEROL, Probable 5-dehydro-4-deoxyglucarate dehydratase | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Structure and function of a decarboxylating Agrobacterium tumefaciens keto-deoxy-d-galactarate dehydratase.
Biochemistry, 53, 2014
|
|
4WBU
 
 | | prion peptide | | Descriptor: | PrP peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
7VRS
 
 | | The complex of Acyltransferase and Acyl Carrier Protein Domains from module 9 of Salinomycin Polyketide Synthase | | Descriptor: | 1,1'-butane-1,4-diylbis(1H-pyrrole-2,5-dione), 4'-PHOSPHOPANTETHEINE, Type I modular polyketide synthase | | Authors: | Feng, Y, Zheng, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural visualization of transient interactions between the cis-acting acyltransferase and acyl carrier protein of the salinomycin modular polyketide synthase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7D5V
 
 | | Structure of the C646A mutant of peptidylarginine deiminase type III (PAD3) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein-arginine deiminase type-3 | | Authors: | Akimoto, M, Mashimo, R, Unno, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
5AP1
 
 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 6-{[3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl]amino}-2-(cyclohexylamino)pyridine-3-carbonitrile, DUAL SPECIFICITY PROTEIN KINASE TTK, GLYCEROL, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.J, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R.L.M, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
9OPV
 
 | | Herpes simplex virus type 1 (HSV-1) C-Capsid portal turrets | | Descriptor: | Capsid portal protein | | Authors: | Crofut, E.H, Kashyap, S, Stevens, A, Jih, J, Liu, Y.-T, Zhou, Z.H. | | Deposit date: | 2025-05-20 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a new capsid form and comparison with A-, B-, and C-capsids clarify herpesvirus assembly.
J.Virol., 99, 2025
|
|
