7DUG
 
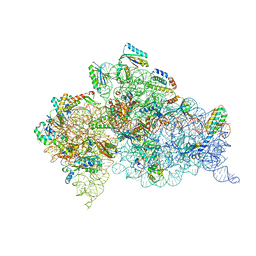 | |
7DUI
 
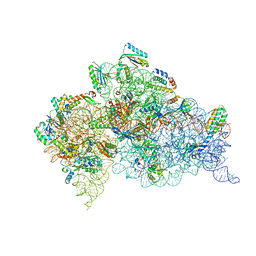 | |
7DUJ
 
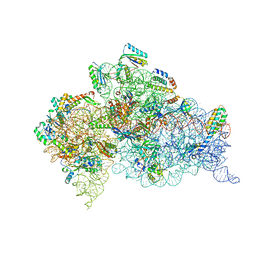 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with mRNA and cognate transfer RNA anticodon stem-loop and sisomicin derivative N1,3''Bz bound | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, 30S Ribosomal RNA rRNA, 30S ribosomal protein S10, ... | | Authors: | DeMirci, H, Destan, E. | | Deposit date: | 2021-01-09 | | Release date: | 2022-01-12 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with mRNA and cognate transfer RNA anticodon stem-loop and sisomicin derivative N1,3''Bz bound
to be published
|
|
8DGJ
 
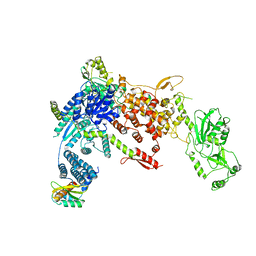 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ib | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DGI
 
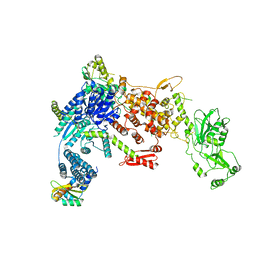 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ia | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
5L2L
 
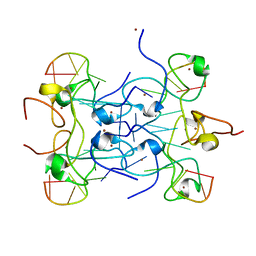 | | Nab2 Zn fingers 5-7 bound to A11G RNA | | Descriptor: | Nab2p, RNA (5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*G)-3'), ZINC ION | | Authors: | Stewart, M, Aibara, S. | | Deposit date: | 2016-08-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the dimerization of Nab2 generated by RNA binding provides insight into its contribution to both poly(A) tail length determination and transcript compaction in Saccharomyces cerevisiae.
Nucleic Acids Res., 45, 2017
|
|
4ZT0
 
 | |
6TFG
 
 | |
6TFF
 
 | |
6D06
 
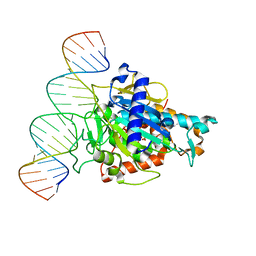 | | Human ADAR2d E488Y mutant complexed with dsRNA containing an abasic site opposite the edited base | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*CP*AP*GP*AP*GP*CP*CP*CP*CP*CP*NP*AP*GP*CP*AP*UP*CP*GP*CP*GP*AP*GP*C)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Bump-Hole Approach for Directed RNA Editing.
Cell Chem Biol, 26, 2019
|
|
4FLA
 
 | | Crystal structure of human RPRD1B, carboxy-terminal domain | | Descriptor: | Regulation of nuclear pre-mRNA domain-containing protein 1B, UNKNOWN ATOM OR ION | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4LQ3
 
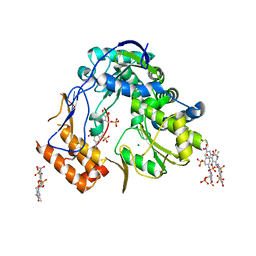 | | Crystal structure of human norovirus RNA-dependent RNA-polymerase bound to the inhibitor PPNDS | | Descriptor: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, 5'-R(P*GP*G)-3', MAGNESIUM ION, ... | | Authors: | Milani, M, Tarantino, D, Mastrangelo, E, Croci, R. | | Deposit date: | 2013-07-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Naphthalene-sulfonate inhibitors of human norovirus RNA-dependent RNA-polymerase.
Antiviral Res., 102, 2014
|
|
8D8N
 
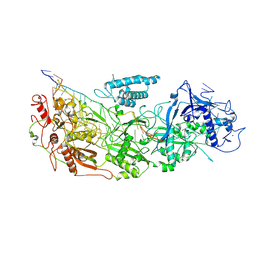 | | gRAMP non-match PFS target RNA | | Descriptor: | RAMP superfamily protein, RNA (35-MER), RNA (5'-R(P*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*AP*CP*A)-3'), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
6BFB
 
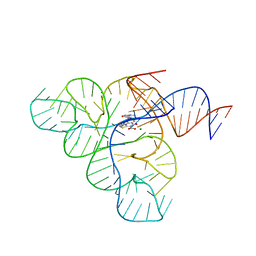 | | Crystal structure of a F. nucleatum FMN riboswitch bound to WG-3 | | Descriptor: | 5-[(3S,4S)-3-(dimethylamino)-4-hydroxypyrrolidin-1-yl]-6-fluoro-4-methyl-8-oxo-3,4-dihydro-8H-1-thia-4,9b-diazacyclopenta[cd]phenalene-9-carboxylic acid, MAGNESIUM ION, RNA (54-MER), ... | | Authors: | Rizvi, N.F, Fischmann, T.O. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of Selective RNA-Binding Small Molecules by Affinity-Selection Mass Spectrometry.
ACS Chem. Biol., 13, 2018
|
|
2PJP
 
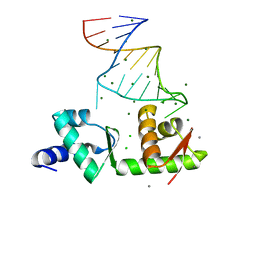 | | Structure of the mRNA-binding domain of elongation factor SelB from E.coli in complex with SECIS RNA | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Soler, N, Fourmy, D, Yoshizawa, S. | | Deposit date: | 2007-04-16 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into a molecular switch in tandem winged-helix motifs from elongation factor SelB.
J.Mol.Biol., 370, 2007
|
|
6T0O
 
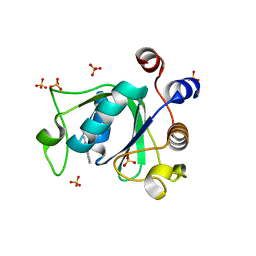 | | Crystal structure of YTHDC1 with fragment 14 (ACA_DC1_004) | | Descriptor: | 2-methyl-3~{H}-pyrido[3,4-d]pyrimidin-4-one, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6SZ8
 
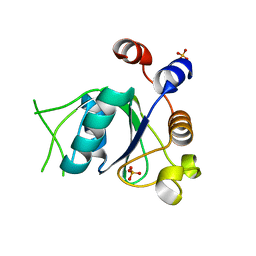 | | Crystal structure of YTHDC1 with fragment 6 (DHU_DC1_034) | | Descriptor: | 6-pyrrolidin-1-yl-5~{H}-pyrimidine-2,4-dione, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6SZN
 
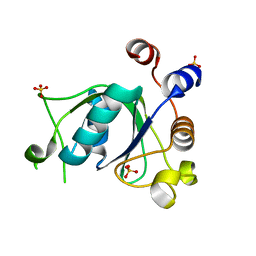 | | Crystal structure of YTHDC1 with fragment 8 (DHU_DC1_006) | | Descriptor: | N-carbamoylbenzamide, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6SZT
 
 | | Crystal structure of YTHDC1 with fragment 10 (DHU_DC1_076) | | Descriptor: | 6-[[methyl(thiophen-3-ylmethyl)amino]methyl]-5~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T02
 
 | | Crystal structure of YTHDC1 with fragment 15 (DHU_DC1_169) | | Descriptor: | (~{S})-phenyl-[(2~{S})-pyrrolidin-2-yl]methanol, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T0Z
 
 | | Crystal structure of YTHDC1 with fragment 23 (ACA_DC1_005) | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-cyclopropyl-1~{H}-imidazole-4-sulfonamide | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T07
 
 | | Crystal structure of YTHDC1 with fragment 20 (DHU_DC1_134) | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N}-[(2~{S})-pyrrolidin-2-yl]-1~{H}-1,2,4-triazol-5-amine | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T0X
 
 | | Crystal structure of YTHDC1 with fragment 22 (ACA_DC1_001) | | Descriptor: | (3~{S})-~{N}-methylpyrrolidine-3-carboxamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T11
 
 | | Crystal structure of YTHDC1 with fragment 29 (DHU_DC1_218) | | Descriptor: | N-methyl-1H-indole-7-carboxamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
4Z7L
 
 | | Crystal structure of Cas6b | | Descriptor: | Cas6b, RNA (5'-R(*GP*CP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*GP*C)-3'), SULFATE ION | | Authors: | Li, H. | | Deposit date: | 2015-04-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | A Non-Stem-Loop CRISPR RNA Is Processed by Dual Binding Cas6.
Structure, 24, 2016
|
|
