2WQT
 
 | | Dodecahedral assembly of MhpD | | Descriptor: | 2-KETO-4-PENTENOATE HYDRATASE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Montgomery, M.G, Wood, S.P. | | Deposit date: | 2009-08-27 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Assembly of a 20Nm Protein Cage by Escherichia Coli 2-Hydroxypentadienoic Acid Hydratase (Mhpd).
J.Mol.Biol., 396, 2010
|
|
2BXP
 
 | | Human serum albumin complexed with myristate and phenylbutazone | | Descriptor: | 4-BUTYL-1,2-DIPHENYL-PYRAZOLIDINE-3,5-DIONE, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
2BXB
 
 | | Human serum albumin complexed with oxyphenbutazone | | Descriptor: | 4-BUTYL-1-(4-HYDROXYPHENYL)-2-PHENYLPYRAZOLIDINE-3,5-DIONE, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
2BXC
 
 | | Human serum albumin complexed with phenylbutazone | | Descriptor: | 4-BUTYL-1,2-DIPHENYL-PYRAZOLIDINE-3,5-DIONE, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
2BXO
 
 | | Human serum albumin complexed with myristate and oxyphenbutazone | | Descriptor: | 4-BUTYL-1-(4-HYDROXYPHENYL)-2-PHENYLPYRAZOLIDINE-3,5-DIONE, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
6QCF
 
 | | Ovine respiratory complex I FRC open class 6 | | Descriptor: | Acyl carrier protein, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
6QC5
 
 | | Ovine respiratory complex I FRC closed class 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-26 | | Release date: | 2019-08-21 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
2BXQ
 
 | | Human serum albumin complexed with myristate, phenylbutazone and indomethacin | | Descriptor: | 4-BUTYL-1,2-DIPHENYL-PYRAZOLIDINE-3,5-DIONE, INDOMETHACIN, MYRISTIC ACID, ... | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
3A68
 
 | | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin | | Descriptor: | ACETIC ACID, CALCIUM ION, Ferritin-4, ... | | Authors: | Masuda, T, Goto, F, Yoshihara, T, Mikami, B. | | Deposit date: | 2009-08-26 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin
J.Biol.Chem., 285, 2010
|
|
2FKW
 
 | | Structure of LH2 from Rps. acidophila crystallized in lipidic mesophases | | Descriptor: | BACTERIOCHLOROPHYLL A, LAURYL DIMETHYLAMINE-N-OXIDE, Light-harvesting protein B-800/850, ... | | Authors: | Papiz, M.Z, Cherezov, V, Clogston, J, Caffrey, M. | | Deposit date: | 2006-01-05 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Room to Move: Crystallizing Membrane Proteins in Swollen Lipidic Mesophases
J.Mol.Biol., 357, 2006
|
|
2AXT
 
 | | Crystal Structure of Photosystem II from Thermosynechococcus elongatus | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Loll, B, Kern, J, Saenger, W, Zouni, A, Biesiadka, J. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-27 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II
NATURE, 438, 2005
|
|
2G50
 
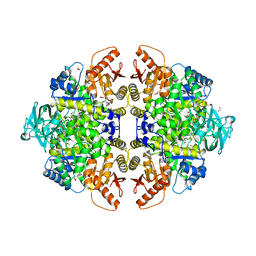 | | The location of the allosteric amino acid binding site of muscle pyruvate kinase. | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ALANINE, ... | | Authors: | Holyoak, T, Williams, R, Fenton, A.W. | | Deposit date: | 2006-02-22 | | Release date: | 2006-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differentiating a Ligand's Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase.
Biochemistry, 45, 2006
|
|
3AG1
 
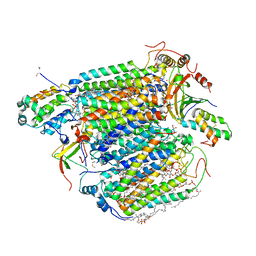 | | Bovine Heart Cytochrome c Oxidase in the Carbon Monoxide-bound Fully Reduced State at 280 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2ZZS
 
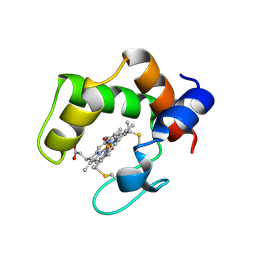 | | Crystal structure of cytochrome c554 from Vibrio parahaemolyticus strain RIMD2210633 | | Descriptor: | Cytochrome c554, GLYCEROL, HEME C | | Authors: | Akazaki, H, Kawai, F, Kumaki, Y, Sekine, K, Hakamata, W, Nishio, T, Park, S.-Y, Oku, T. | | Deposit date: | 2009-02-24 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cytochrome c554 from Vibrio parahaemolyticus strain RIMD2210633
To be Published
|
|
3AG4
 
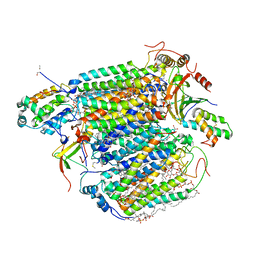 | | Bovine Heart Cytochrome c Oxidase in the Cyanide Ion-bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2HH0
 
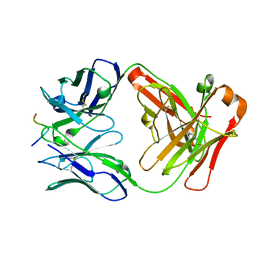 | |
3A0B
 
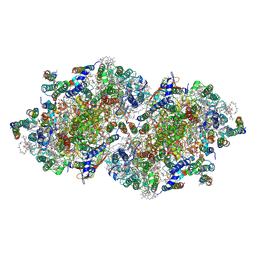 | | Crystal structure of Br-substituted Photosystem II complex | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Kawakami, K, Umena, Y, Kamiya, N, Shen, J.-R. | | Deposit date: | 2009-03-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ABM
 
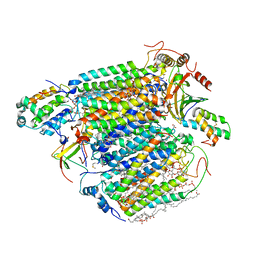 | | Bovine heart cytochrome c oxidase at the fully oxidized state (200-s X-ray exposure dataset) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Aoyama, H, Muramoto, K, Shinzawa-Itoh, K, Yamashita, E, Tsukihara, T, Ogura, T, Yoshikawa, S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A peroxide bridge between Fe and Cu ions in the O2 reduction site of fully oxidized cytochrome c oxidase could suppress the proton pump
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1Z8G
 
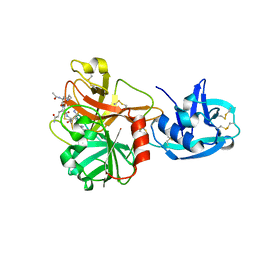 | | Crystal structure of the extracellular region of the transmembrane serine protease hepsin with covalently bound preferred substrate. | | Descriptor: | ACE-LYS-GLN-LEU-ARG-Chloromethylketone, Serine protease hepsin | | Authors: | Herter, S, Piper, D.E, Aaron, W, Gabriele, T, Cutler, G, Cao, P, Bhatt, A.S, Choe, Y, Craik, C.S, Walker, N, Meininger, D, Hoey, T, Austin, R.J. | | Deposit date: | 2005-03-30 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membrane-anchored serine protease implicated in prostate and ovarian cancers
Biochem.J., 390, 2005
|
|
2DCN
 
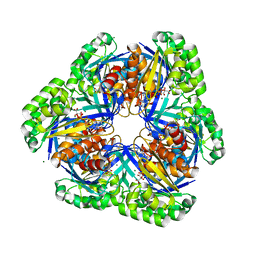 | | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate (alpha-furanose form) | | Descriptor: | 6-O-phosphono-beta-D-psicofuranosonic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Okazaki, S, Onda, H, Suzuki, A, Kuramitsu, S, Masui, R, Yamane, T. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate
To be Published
|
|
2DYS
 
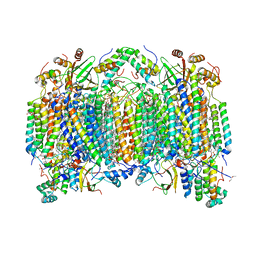 | | Bovine heart cytochrome C oxidase modified by DCCD | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shinzawa-Itoh, K, Aoyama, H, Muramoto, K, Kurauchi, T, Mizushima, T, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2006-09-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase
Embo J., 26, 2007
|
|
2EIN
 
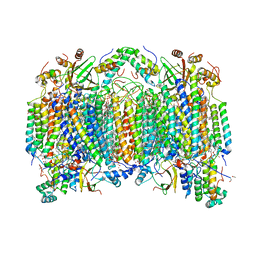 | | Zinc ion binding structure of bovine heart cytochrome C oxidase in the fully oxidized state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Hirata, K, Shinzawa-Itoh, K, Yoko-o, S, Yamashita, E, Aoyama, H, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2007-03-13 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A histidine residue acting as a controlling site for dioxygen reduction and proton pumping by cytochrome c oxidase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7X76
 
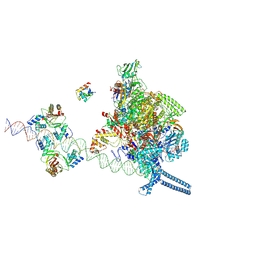 | |
7X74
 
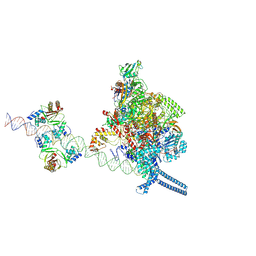 | |
7X75
 
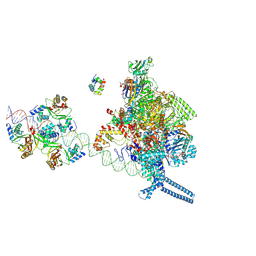 | |
