1R1R
 
 | |
6LDY
 
 | |
5JLR
 
 | | Crystal structure of Mycobacterium avium SerB2 with serine present at slightly different position near ACT domain | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, MAGNESIUM ION, ... | | Authors: | Shree, S, Agrawal, A, Dubey, S, Ramachandran, R. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Crystal structure of Mycobacterium avium SerB2 with serine present at slightly different position near ACT domain
To be published
|
|
1OGH
 
 | |
6LDW
 
 | |
5IWC
 
 | | Mycobacterium tuberculosis CysM in complex with the Urea-scaffold inhibitor 3 [4-(3-([1,1'-Biphenyl]-3-yl)ureido)-2-hydroxybenzoic acid] | | Descriptor: | 4-{[([1,1'-biphenyl]-3-yl)carbamoyl]amino}-2-hydroxybenzoic acid, O-phosphoserine sulfhydrylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schnell, R, Maric, S, Lindqvist, Y, Schneider, G. | | Deposit date: | 2016-03-22 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis.
J.Med.Chem., 59, 2016
|
|
1P63
 
 | | Human Acidic Fibroblast Growth Factor. 140 Amino Acid Form with Amino Terminal His Tag and Leu111 Replaced with Ile (L111I) | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR, FORMIC ACID, SULFATE ION | | Authors: | Brych, S.R, Kim, J, Logan, T.M, Blaber, M. | | Deposit date: | 2003-04-28 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein
superfold
Protein Sci., 12, 2003
|
|
6LDV
 
 | |
1Q9G
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX
J.Mol.Biol., 336, 2004
|
|
6M2V
 
 | | Crystal structure of UHRF1 SRA complexed with fully-mCHG DNA. | | Descriptor: | DNA (5'-D(*TP*CP*AP*CP*GP*(5CM)P*TP*GP*CP*GP*TP*GP*A)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Abhishek, S, Nakarakanti, N.K, Deeksha, W, Rajakumara, E. | | Deposit date: | 2020-03-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic insights into recognition of symmetric methylated cytosines in CpG and non-CpG DNA by UHRF1 SRA.
Int.J.Biol.Macromol., 170, 2021
|
|
1QPF
 
 | | FK506 BINDING PROTEIN (12 KDA, HUMAN) COMPLEX WITH L-709,858 | | Descriptor: | C32-O-(1-ETHYL-INDOL-5-YL)ASCOMYCIN, PROTEIN (FK506-BINDING PROTEIN), heptyl beta-D-glucopyranoside | | Authors: | Becker, J.W, Rotonda, J. | | Deposit date: | 1999-05-24 | | Release date: | 1999-08-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 32-Indolyl ether derivatives of ascomycin: three-dimensional structures of complexes with FK506-binding protein.
J.Med.Chem., 42, 1999
|
|
5JJB
 
 | |
5JMA
 
 | | Crystal structure of Mycobacterium avium SerB2 in complex with serine at catalytic (PSP) domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Phosphoserine phosphatase, ... | | Authors: | Shree, S, Dubey, S, Agrawal, A, Ramachandran, R. | | Deposit date: | 2016-04-28 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Mycobacterium avium SerB2 in complex with serine at catalytic (PSP) domain
To be published
|
|
6LRB
 
 | |
1PMV
 
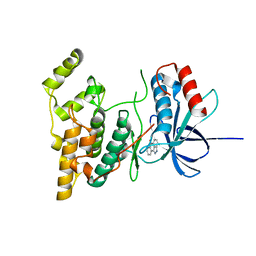 | | The structure of JNK3 in complex with a dihydroanthrapyrazole inhibitor | | Descriptor: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, Mitogen-activated protein kinase 10 | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, LoGrasso, P.V. | | Deposit date: | 2003-06-11 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity
Chem.Biol., 10, 2003
|
|
6LT0
 
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | Descriptor: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | Authors: | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1PS7
 
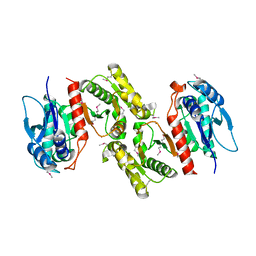 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
5IT0
 
 | |
1PS6
 
 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-HYDROXY-L-THREONINE-5-MONOPHOSPHATE, 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1Q2N
 
 | |
6M4O
 
 | | Cryo-EM structure of the monomeric SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-03-08 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6M4N
 
 | | Cryo-EM structure of the dimeric SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-03-07 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1RTT
 
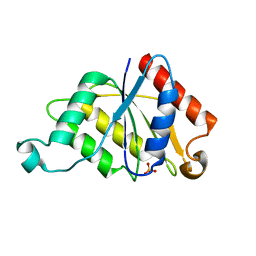 | | Crystal structure determination of a putative NADH-dependent reductase using sulfur anomalous signal | | Descriptor: | SULFATE ION, conserved hypothetical protein | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure determination of an FMN reductase from Pseudomonas aeruginosa PA01 using sulfur anomalous signal.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
5JLP
 
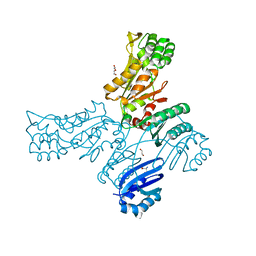 | |
1RQ1
 
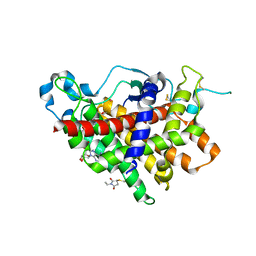 | | Structure of Ero1p, Source of Disulfide Bonds for Oxidative Protein Folding in the Cell | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, CADMIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Gross, E, Kastner, D.B, Kaiser, C.A, Fass, D. | | Deposit date: | 2003-12-04 | | Release date: | 2004-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of ero1p, source of disulfide bonds for oxidative protein folding in the cell.
Cell(Cambridge,Mass.), 117, 2004
|
|
