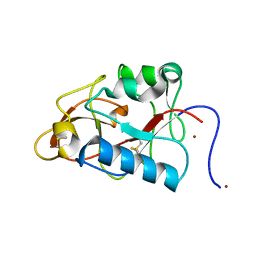
1DY0
 
 | |
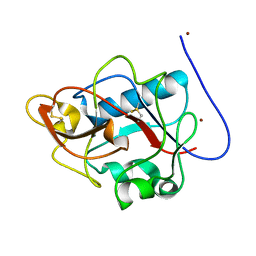
1DY1
 
 | |
1DY2
 
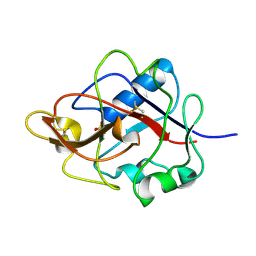 | | Murine collagen alpha1(XV), endostatin domain | | Descriptor: | COLLAGEN ALPHA1(XV) CHAIN, SULFATE ION | | Authors: | Tisi, D, Hohenester, E, Sasaki, T, Timpl, R. | | Deposit date: | 2000-01-21 | | Release date: | 2001-01-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Endostatin Derived from Collagens Xv and Xviii Differ in Structural and Binding Properties, Tissue Distribution and Anti-Angiogenic Activity
J.Mol.Biol., 301, 2000
|
|
1DY3
 
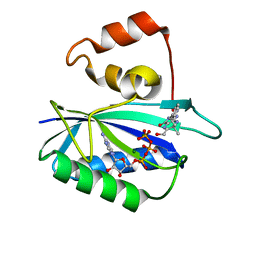 | | Ternary complex of 7,8-dihydro-6-hydroxymethylpterinpyrophosphokinase from Escherichia coli with ATP and a substrate analogue. | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 7,8-DIHYDRO-6-HYDROXYMETHYL-7-METHYL-7-[2-PHENYLETHYL]-PTERIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stammers, D.K, Achari, A, Somers, D.O, Bryant, P.K, Rosemond, J, Scott, D.L, Champness, J.N. | | Deposit date: | 2000-01-21 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A X-Ray Structure of the Ternary Complex of 7,8-Dihydro-6-Hydroxymethylpterinpyrophosphokinase from Escherichia Coli with ATP and a Substrate Analogue
FEBS Lett., 456, 1999
|
|
1DY4
 
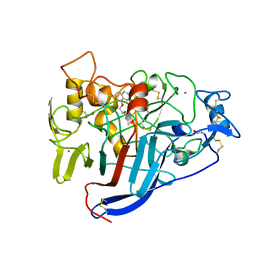 | | CBH1 IN COMPLEX WITH S-PROPRANOLOL | | Descriptor: | 1-(ISOPROPYLAMINO)-3-(1-NAPHTHYLOXY)-2-PROPANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Stahlberg, J, Henriksson, H, Divne, C, Isaksson, R, Pettersson, G, Johansson, G, Jones, T.A. | | Deposit date: | 2000-01-26 | | Release date: | 2000-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Enantiomer Binding and Separation of a Common Beta-Blocker: Crystal Structure of Cellobiohydrolase Cel7A with Bound (S)-Propranolol at 1.9 A Resolution
J.Mol.Biol., 305, 2001
|
|
1DY5
 
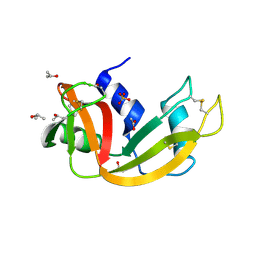 | | Deamidated derivative of bovine pancreatic ribonuclease | | Descriptor: | ACETATE ION, ISOPROPYL ALCOHOL, RIBONUCLEASE A, ... | | Authors: | Esposito, L, Vitagliano, L, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 2000-01-27 | | Release date: | 2000-03-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | The Ultrahigh Resolution Crystal Structure of Ribonuclease A Containing an Isoaspartyl Residue: Hydration and Sterochemical Analysis.
J.Mol.Biol., 297, 2000
|
|
1DY6
 
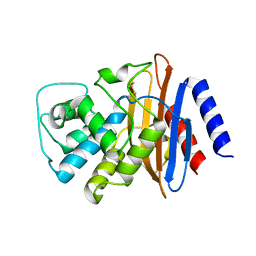 | | Structure of the imipenem-hydrolyzing beta-lactamase SME-1 | | Descriptor: | CARBAPENEM-HYDROLYSING BETA-LACTAMASE SME-1 | | Authors: | Sougakoff, W, L'Hermite, G, Billy, I, Guillet, V, Naas, T, Nordman, P, Jarlier, V, Delettre, J. | | Deposit date: | 2000-01-27 | | Release date: | 2001-01-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of the Imipenem-Hydrolyzing Class a Beta-Lactamase Sme-1 from Serratia Marcescens.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1DY7
 
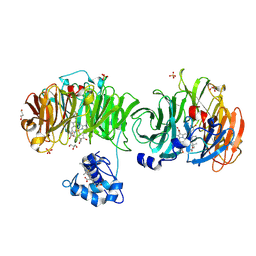 | | Cytochrome cd1 Nitrite Reductase, CO complex | | Descriptor: | CARBON MONOXIDE, GLYCEROL, HEME C, ... | | Authors: | Sjogren, T, Svensson-Ek, M, Hajdu, J, Brzezinski, P. | | Deposit date: | 2000-01-28 | | Release date: | 2000-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proton-Coupled Structural Changes Upon Binding of Carbon Monoxide to Cytochrome Cd(1): A Combined Flash Photolysis and X-Ray Crystallography Study
Biochemistry, 39, 2000
|
|
1DY8
 
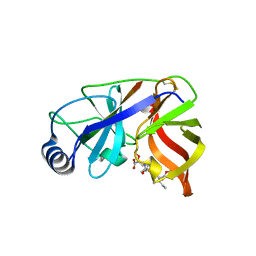 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor II) | | Descriptor: | N-[(benzyloxy)carbonyl]-L-isoleucyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70) | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1DY9
 
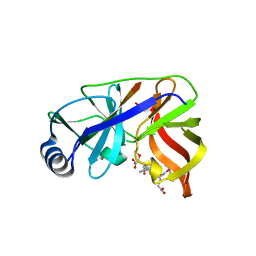 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor I) | | Descriptor: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1DYA
 
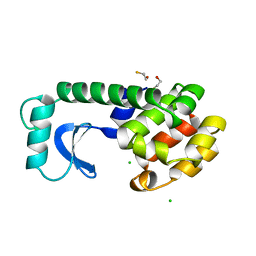 | |
1DYB
 
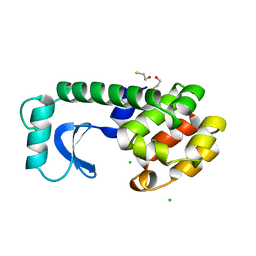 | |
1DYC
 
 | |
1DYD
 
 | |
1DYE
 
 | |
1DYF
 
 | |
1DYG
 
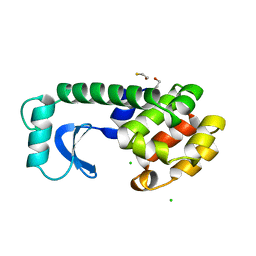 | |
1DYH
 
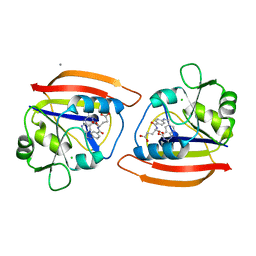 | | ISOMORPHOUS CRYSTAL STRUCTURES OF ESCHERICHIA COLI DIHYDROFOLATE REDUCTASE COMPLEXED WITH FOLATE, 5-DEAZAFOLATE AND 5,10-DIDEAZATETRAHYDROFOLATE: MECHANISTIC IMPLICATIONS | | Descriptor: | 5-DEAZAFOLIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reyes, V.M, Kraut, J. | | Deposit date: | 1994-10-26 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Isomorphous crystal structures of Escherichia coli dihydrofolate reductase complexed with folate, 5-deazafolate, and 5,10-dideazatetrahydrofolate: mechanistic implications.
Biochemistry, 34, 1995
|
|
1DYI
 
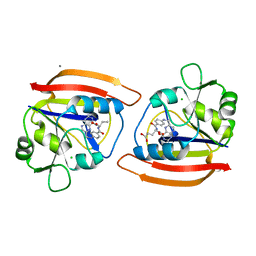 | | ISOMORPHOUS CRYSTAL STRUCTURES OF ESCHERICHIA COLI DIHYDROFOLATE REDUCTASE COMPLEXED WITH FOLATE, 5-DEAZAFOLATE AND 5,10-DIDEAZATETRAHYDROFOLATE: MECHANISTIC IMPLICATIONS | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Reyes, V.M, Kraut, J. | | Deposit date: | 1994-10-26 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Isomorphous crystal structures of Escherichia coli dihydrofolate reductase complexed with folate, 5-deazafolate, and 5,10-dideazatetrahydrofolate: mechanistic implications.
Biochemistry, 34, 1995
|
|
1DYJ
 
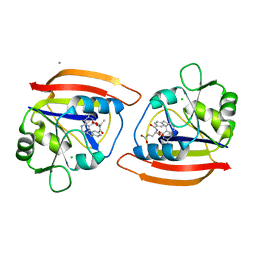 | | ISOMORPHOUS CRYSTAL STRUCTURES OF ESCHERICHIA COLI DIHYDROFOLATE REDUCTASE COMPLEXED WITH FOLATE, 5-DEAZAFOLATE AND 5,10-DIDEAZATETRAHYDROFOLATE: MECHANISTIC IMPLICATIONS | | Descriptor: | 5,10-DIDEAZATETRAHYDROFOLIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reyes, V.M, Kraut, J. | | Deposit date: | 1994-10-26 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Isomorphous crystal structures of Escherichia coli dihydrofolate reductase complexed with folate, 5-deazafolate, and 5,10-dideazatetrahydrofolate: mechanistic implications.
Biochemistry, 34, 1995
|
|
1DYK
 
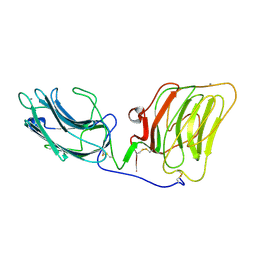 | | Laminin alpha 2 chain LG4-5 domain pair | | Descriptor: | CALCIUM ION, LAMININ ALPHA 2 CHAIN | | Authors: | Tisi, D, Talts, J.F, Timple, R, Hohenester, E. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the C-Terminal Laminin G-Like Domain Pair of the Laminin Alpha 2 Chain Harbouring Binding Sites for Alpha-Dystroglycan and Heparin
Embo J., 19, 2000
|
|
1DYL
 
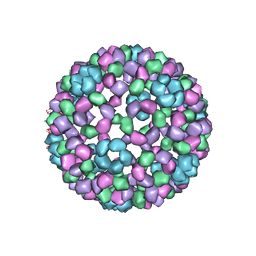 | | 9 ANGSTROM RESOLUTION CRYO-EM RECONSTRUCTION STRUCTURE OF SEMLIKI FOREST VIRUS (SFV) AND FITTING OF THE CAPSID PROTEIN STRUCTURE IN THE EM DENSITY | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Mancini, E.J, Clarke, M, Gowen, B.E, Rutten, T, Fuller, S.D. | | Deposit date: | 2000-02-02 | | Release date: | 2000-08-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Electron Microscopy Reveals the Functional Organization of an Enveloped Virus, Semliki Forest Virus.
Mol.Cell, 5, 2000
|
|
1DYM
 
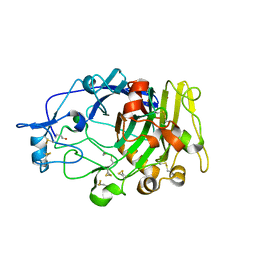 | | Humicola insolens Endocellulase Cel7B (EG 1) E197A Mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Moraz, O, Driguez, H, Schulein, M. | | Deposit date: | 2000-02-03 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335 ( Pt 2), 1998
|
|
1DYN
 
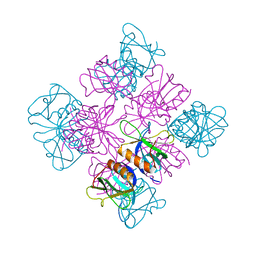 | | CRYSTAL STRUCTURE AT 2.2 ANGSTROMS RESOLUTION OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM HUMAN DYNAMIN | | Descriptor: | DYNAMIN | | Authors: | Ferguson, K.M, Lemmon, M.A, Schlessinger, J, Sigler, P.B. | | Deposit date: | 1994-12-21 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin.
Cell(Cambridge,Mass.), 79, 1994
|
|
1DYO
 
 | | Xylan-Binding Domain from CBM 22, formally x6b domain | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Davies, G.J, Charnock, S.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2000-02-03 | | Release date: | 2000-07-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X6 Thermostabilising Domains of Xylanases are Carbohydrate Binding Modules: Structure and Biochemistry of the Clostridium Thermocellum X6B Domain
Biochemistry, 39, 2000
|
|
