1KLA
 
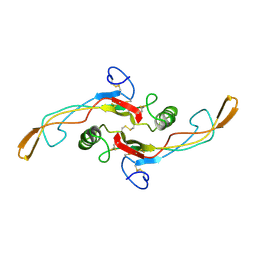 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 1-17 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1LRE
 
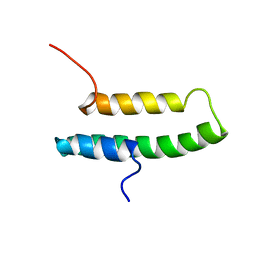 | |
1LIR
 
 | | LQ2 FROM LEIURUS QUINQUESTRIATUS, NMR, 22 STRUCTURES | | Descriptor: | LQ2 | | Authors: | Renisio, J.G, Lu, Z, Blanc, E, Jin, W, Lewis, J.H, Bornet, O, Darbon, H. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of potassium channel-inhibiting scorpion toxin Lq2.
Proteins, 34, 1999
|
|
1ZDM
 
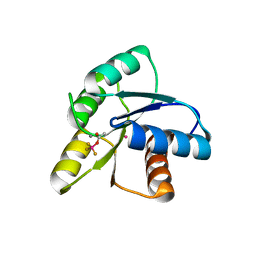 | | Crystal Structure of Activated CheY Bound to Xe | | Descriptor: | Chemotaxis protein cheY, MANGANESE (II) ION, XENON | | Authors: | Lowery, T.J, Doucleff, M, Ruiz, E.J, Rubin, S.M, Pines, A, Wemmer, D.E. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinguishing multiple chemotaxis Y protein conformations with laser-polarized 129Xe NMR.
Protein Sci., 14, 2005
|
|
1L4T
 
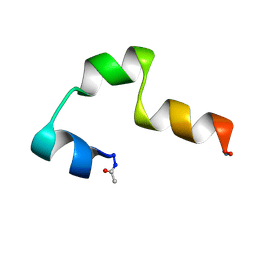 | | SOLUTION NMR STRUCTURE OF THE CCK2E3 | | Descriptor: | Gastrin/cholecystokinin type B receptor | | Authors: | Giragossian, C, Mierke, D.F. | | Deposit date: | 2002-03-05 | | Release date: | 2003-01-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Intermolecular interactions between cholecystokinin-8 and the third extracellular loop of the cholecystokinin-2 receptor
Biochemistry, 41, 2002
|
|
1Y9O
 
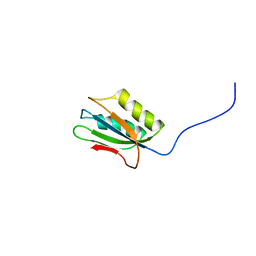 | | 1H NMR Structure of Acylphosphatase from the hyperthermophile Sulfolobus Solfataricus | | Descriptor: | Acylphosphatase | | Authors: | Corazza, A, Rosano, C, Pagano, K, Alverdi, V, Esposito, G, Capanni, C, Bemporad, F, Plakoutsi, G, Stefani, M, Chiti, F, Zuccotti, S, Bolognesi, M, Viglino, P. | | Deposit date: | 2004-12-16 | | Release date: | 2005-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, conformational stability, and enzymatic properties of acylphosphatase from the hyperthermophile Sulfolobus solfataricus
Proteins, 62, 2006
|
|
1ZZA
 
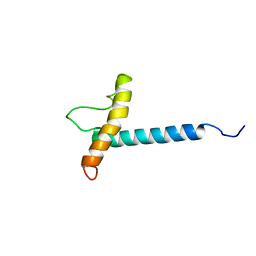 | | Solution NMR Structure of the Membrane Protein Stannin | | Descriptor: | Stannin | | Authors: | Buck-Koehntop, B.A, Mascioni, A, Buffy, J.J, Veglia, G. | | Deposit date: | 2005-06-13 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and membrane topology of stannin: a mediator of neuronal cell apoptosis induced by trimethyltin chloride.
J.Mol.Biol., 354, 2005
|
|
1JVR
 
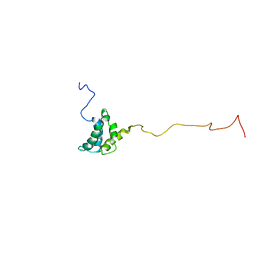 | | STRUCTURE OF THE HTLV-II MATRIX PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | HUMAN T-CELL LEUKEMIA VIRUS TYPE II MATRIX PROTEIN | | Authors: | Christensen, A.M, Massiah, M.A, Turner, B.G, Sundquist, W.I, Summers, M.F. | | Deposit date: | 1996-10-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses.
J.Mol.Biol., 264, 1996
|
|
1LIP
 
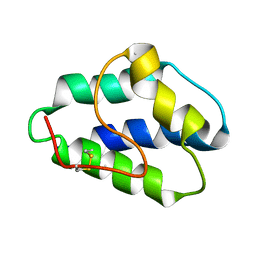 | |
2L8S
 
 | |
2AKG
 
 | |
2NDE
 
 | | Solution Structure of Mutant of BMAP-28(1-18) | | Descriptor: | Cathelicidin-5 | | Authors: | Agadi, N, Kumar, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Function And Membrane Interaction Studies of Two Synthetic Peptides Using Solution And Solid State NMR
To be Published
|
|
2NDC
 
 | | Solution Structure of BMAP-28(1-18) | | Descriptor: | Cathelicidin-5 | | Authors: | Agadi, N, Kumar, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Function And Membrane Interaction Studies Of Two Synthetic Antimicrobial Peptides Using Solution And Solid State NMR
To be Published
|
|
1WPI
 
 | | Solution NMR Structure of Protein YKR049C from Saccharomyces cerevisiae. Ontario Centre for Structural Proteomics target YST0250_1_133; Northeast Structural Genomics Consortium YTYst250 | | Descriptor: | Hypothetical 15.6 kDa protein in NAP1-TRK2 intergenic region | | Authors: | Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-03 | | Release date: | 2005-09-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of YKR049C, a putative redox protein from Saccharomyces cerevisiae
J.Biochem.Mol.Biol., 38, 2005
|
|
2EOT
 
 | | SOLUTION STRUCTURE OF EOTAXIN, AN ENSEMBLE OF 32 NMR SOLUTION STRUCTURES | | Descriptor: | EOTAXIN | | Authors: | Crump, M.P, Rajarathnam, K, Kim, K.-S, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 1998-06-29 | | Release date: | 1998-11-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of eotaxin, a chemokine that selectively recruits eosinophils in allergic inflammation.
J.Biol.Chem., 273, 1998
|
|
2M54
 
 | | Refined NMR solution structure of metal-modified DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*TP*TP*TP*(D33)P*(D33)P*(D33)P*AP*AP*AP*TP*TP*AP*A)-3'), SILVER ION | | Authors: | Kumbhar, S, Johannsen, S, Sigel, R.K, Waller, M.P, Mueller, J. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A QM/MM refinement of an experimental DNA structure with metal-mediated base pairs.
J.Inorg.Biochem., 127, 2013
|
|
1ZBJ
 
 | |
2LS6
 
 | | Solution NMR Structure of a Non-canonical galactose-binding CBM32 from Clostridium perfringens | | Descriptor: | Hyaluronoglucosaminidase | | Authors: | Grondin, J.M, Chitayat, S, Ficko-Blean, E, Boraston, A.B, Smith, S.P. | | Deposit date: | 2012-04-20 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An unusual mode of galactose recognition by a family 32 carbohydrate-binding module.
J.Mol.Biol., 426, 2014
|
|
2L1A
 
 | | Solution NMR structure of the N-terminal GTPase-like domain of dictyostelium discoideum Fomin C | | Descriptor: | Formin-C | | Authors: | Dames, S.A, Schoenichen, A, Stephan, G, Geyer, M. | | Deposit date: | 2010-07-27 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, lipid binding, and physiological relevance of the putative GTPase-binding domain of Dictyostelium formin C.
J.Biol.Chem., 286, 2011
|
|
1SXM
 
 | | SCORPION TOXIN (NOXIUSTOXIN) WITH HIGH AFFINITY FOR VOLTAGE DEPENDENT POTASSIUM CHANNEL AND LOW AFFINITY FOR CALCIUM DEPENDENT POTASSIUM CHANNEL (NMR AT 20 DEGREES, PH3.5, 39 STRUCTURES) | | Descriptor: | NOXIUSTOXIN | | Authors: | Dauplais, M, Gilquin, B, Possani, L.D, Gurrola-Briones, G, Roumestand, C, Menez, A. | | Deposit date: | 1995-09-07 | | Release date: | 1996-01-29 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional solution structure of noxiustoxin: analysis of structural differences with related short-chain scorpion toxins.
Biochemistry, 34, 1995
|
|
1UG4
 
 | |
1SYZ
 
 | |
2E2Z
 
 | | Solution NMR structure of yeast Tim15, co-chaperone of mitochondrial Hsp70 | | Descriptor: | Tim15, ZINC ION | | Authors: | Momose, T, Ohshima, C, Maeda, M, Endo, T. | | Deposit date: | 2006-11-19 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis of functional cooperation of Tim15/Zim17 with yeast mitochondrial Hsp70
Embo Rep., 8, 2007
|
|
1V28
 
 | | Solution structure of paralytic peptide of the wild Silkmoth, Antheraea yamamai | | Descriptor: | Paralytic Peptide | | Authors: | Kawaguchi, K, Ying, A, Suzuki, K, Kumaki, Y, Demura, M, Nitta, K. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of paralytic peptide of the wild Silkmoth Antheraea yamamai by NMR
To be Published
|
|
1ROD
 
 | | CHIMERIC PROTEIN OF INTERLEUKIN 8 AND HUMAN MELANOMA GROWTH STIMULATING ACTIVITY PROTEIN, NMR | | Descriptor: | CHIMERIC PROTEIN OF INTERLEUKIN 8 AND HUMAN MELANOMA GROWTH STIMULATING ACTIVITY PROTEIN | | Authors: | Roesch, P, Sticht, H, Auer, M, Schmitt, B, Besemer, J, Horcher, M, Kirsch, T, Lindley, I.J.D. | | Deposit date: | 1995-11-24 | | Release date: | 1996-06-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and activity of a chimeric interleukin-8-melanoma-growth-stimulatory-activity protein.
Eur.J.Biochem., 235, 1996
|
|
