6W3M
 
 | |
1QND
 
 | | STEROL CARRIER PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Lopez-Garcia, F, Szyperski, T, Dyer, J.H, Choinowski, T, Seedorf, U, Hauser, H, Wuthrich, K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-07-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Sterol Carrier Protein-2: Implications for the Biological Role
J.Mol.Biol., 295, 2000
|
|
2N7M
 
 | | NMR-SAXS/WAXS Structure of the core of the U4/U6 di-snRNA | | Descriptor: | RNA (92-MER) | | Authors: | Cornilescu, G, Didychuk, A.L, Rodgers, M.L, Michael, L.A, Burke, J.E, Montemayor, E.J, Hoskins, A.A, Butcher, S.E, Tonelli, M. | | Deposit date: | 2015-09-14 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of Multi-Helical RNAs by NMR-SAXS/WAXS: Application to the U4/U6 di-snRNA.
J.Mol.Biol., 428, 2016
|
|
1LQQ
 
 | | ANTI-MAMMAL AND ANTI-INSECT LQQIII SCORPION TOXIN, NMR, 15 STRUCTURES | | Descriptor: | LQQIII | | Authors: | Landon, C, Sodano, P, Vovelle, F, Ptak, M. | | Deposit date: | 1997-05-18 | | Release date: | 1998-05-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | 1H-NMR-derived secondary structure and the overall fold of the potent anti-mammal and anti-insect toxin III from the scorpion Leiurus quinquestriatus quinquestriatus.
Eur.J.Biochem., 236, 1996
|
|
2SOB
 
 | | SN-OB, OB-FOLD SUB-DOMAIN OF STAPHYLOCOCCAL NUCLEASE, NMR, 10 STRUCTURES | | Descriptor: | STAPHYLOCOCCAL NUCLEASE | | Authors: | Alexandrescu, A.T, Gittis, A.G, Abeygunawardana, C, Shortle, D. | | Deposit date: | 1995-09-15 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a stable "OB-fold" sub-domain isolated from staphylococcal nuclease.
J.Mol.Biol., 250, 1995
|
|
1AQ5
 
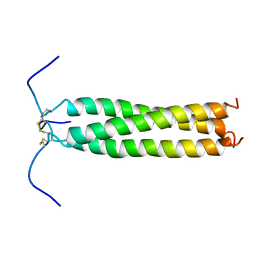 | | HIGH-RESOLUTION SOLUTION NMR STRUCTURE OF THE TRIMERIC COILED-COIL DOMAIN OF CHICKEN CARTILAGE MATRIX PROTEIN, 20 STRUCTURES | | Descriptor: | CARTILAGE MATRIX PROTEIN | | Authors: | Dames, S.A, Wiltscheck, R, Kammerer, R.A, Engel, J, Alexandrescu, A.T. | | Deposit date: | 1997-08-07 | | Release date: | 1998-02-11 | | Last modified: | 2021-01-13 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a parallel homotrimeric coiled coil.
Nat.Struct.Biol., 5, 1998
|
|
1B5K
 
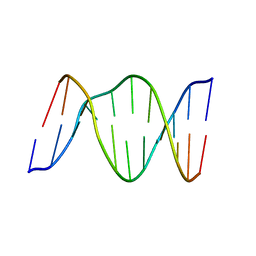 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE THYMIDINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*EDCP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Cullinan, D, Korobka, A, Grollman, A.P, Patel, D.J, Eisenberg, M, De Santos, C.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite thymidine: comparison with the duplex containing deoxyadenosine opposite the adduct.
Biochemistry, 35, 1996
|
|
1BAH
 
 | | A TWO DISULFIDE DERIVATIVE OF CHARYBDOTOXIN WITH DISULFIDE 13-33 REPLACED BY TWO ALPHA-AMINOBUTYRIC ACIDS, NMR, 30 STRUCTURES | | Descriptor: | CHARYBDOTOXIN | | Authors: | Song, J, Gilquin, B, Jamin, N, Guenneugues, M, Dauplais, M, Vita, C, Menez, A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a two-disulfide derivative of charybdotoxin: structural evidence for conservation of scorpion toxin alpha/beta motif and its hydrophobic side chain packing.
Biochemistry, 36, 1997
|
|
1BHU
 
 | | THE 3D STRUCTURE OF THE STREPTOMYCES METALLOPROTEINASE INHIBITOR, SMPI, ISOLATED FROM STREPTOMYCES NIGRESCENS TK-23, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | METALLOPROTEINASE INHIBITOR | | Authors: | Tate, S, Ohno, A, Seeram, S.S, Hiraga, K, Oda, K, Kainosho, M. | | Deposit date: | 1998-06-10 | | Release date: | 1999-01-06 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Streptomyces metalloproteinase inhibitor, SMPI, isolated from Streptomyces nigrescens TK-23: another example of an ancestral beta gamma-crystallin precursor structure.
J.Mol.Biol., 282, 1998
|
|
1BUY
 
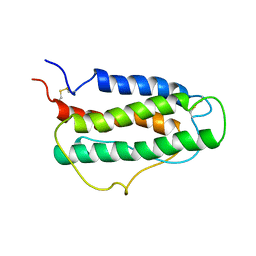 | | HUMAN ERYTHROPOIETIN, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN (ERYTHROPOIETIN) | | Authors: | Cheetham, J.C, Smith, D.M, Aoki, K.H, Stevenson, J.L, Hoeffel, T.J, Syed, R.S, Egrie, J, Harvey, T.S. | | Deposit date: | 1998-09-08 | | Release date: | 1999-09-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of human erythropoietin and a comparison with its receptor bound conformation.
Nat.Struct.Biol., 5, 1998
|
|
1AG2
 
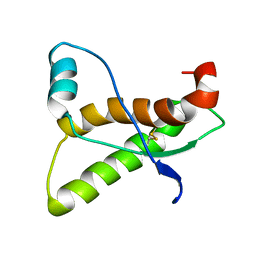 | | PRION PROTEIN DOMAIN PRP(121-231) FROM MOUSE, NMR, 2 MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Billeter, M, Riek, R, Wider, G, Wuthrich, K, Hornemann, S, Glockshuber, R. | | Deposit date: | 1997-03-31 | | Release date: | 1997-10-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the mouse prion protein domain PrP(121-231).
Nature, 382, 1996
|
|
1CUR
 
 | | REDUCED RUSTICYANIN, NMR | | Descriptor: | COPPER (II) ION, CU(I) RUSTICYANIN | | Authors: | Botuyan, M.V, Dyson, H.J. | | Deposit date: | 1996-04-19 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Cu(I) rusticyanin from Thiobacillus ferrooxidans: structural basis for the extreme acid stability and redox potential.
J.Mol.Biol., 263, 1996
|
|
1SYM
 
 | |
1BGZ
 
 | |
1PON
 
 | |
17RA
 
 | |
2N51
 
 | |
1NGR
 
 | |
1T0W
 
 | | 25 NMR structures of Truncated Hevein of 32 aa (Hevein-32) complex with N,N,N-triacetylglucosamina | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevein | | Authors: | Aboitiz, N, Vila-Perello, M, Groves, P, Asensio, J.L, Andreu, D, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2004-04-13 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | NMR and modeling studies of protein-carbohydrate interactions: synthesis, three-dimensional structure, and recognition properties of a minimum hevein domain with binding affinity for chitooligosaccharides
Chembiochem, 5, 2004
|
|
1FSP
 
 | | NMR SOLUTION STRUCTURE OF BACILLUS SUBTILIS SPO0F PROTEIN, 20 STRUCTURES | | Descriptor: | STAGE 0 SPORULATION PROTEIN F | | Authors: | Feher, V.A, Skelton, N.J, Dahlquist, F.W, Cavanagh, J. | | Deposit date: | 1997-06-05 | | Release date: | 1997-12-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition.
Biochemistry, 36, 1997
|
|
1HO9
 
 | |
1DEP
 
 | | MEMBRANE PROTEIN, NMR, 1 STRUCTURE | | Descriptor: | T345-359 | | Authors: | Jung, H, Schnackerz, K.D. | | Deposit date: | 1995-08-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR and circular dichroism studies of synthetic peptides derived from the third intracellular loop of the beta-adrenoceptor.
FEBS Lett., 358, 1995
|
|
1DDF
 
 | | FAS DEATH DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FAS | | Authors: | Huang, B, Eberstadt, M, Olejniczak, E, Meadows, R.P, Fesik, S. | | Deposit date: | 1996-11-08 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain.
Nature, 384, 1996
|
|
1CFE
 
 | | P14A, NMR, 20 STRUCTURES | | Descriptor: | PATHOGENESIS-RELATED PROTEIN P14A | | Authors: | Fernandez, C, Szyperski, T, Bruyere, T, Ramage, P, Mosinger, E, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the pathogenesis-related protein P14a.
J.Mol.Biol., 266, 1997
|
|
1KMR
 
 | |
