7RC5
 
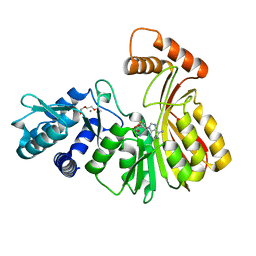 | | Aeronamide N-methyltransferase, AerE (N231A) | | Descriptor: | CALCIUM ION, Methyltransferase family protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC2
 
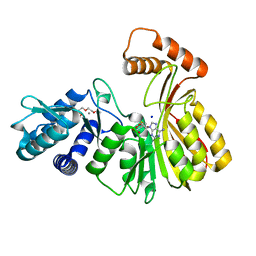 | | Aeronamide N-methyltransferase, AerE | | Descriptor: | CALCIUM ION, Methyltransferase family protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1V8J
 
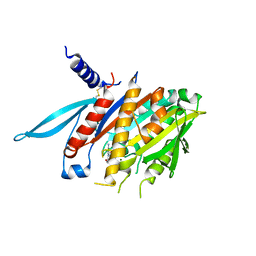 | | The Crystal Structure of the Minimal Functional Domain of the Microtubule Destabilizer KIF2C Complexed with Mg-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF2C, MAGNESIUM ION | | Authors: | Ogawa, T, Nitta, R, Okada, Y, Hirokawa, N. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops.
Cell(Cambridge,Mass.), 116, 2004
|
|
7K5W
 
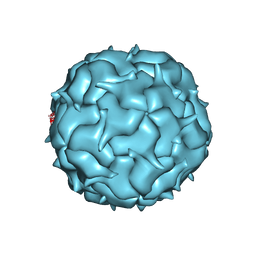 | | Cryo-EM structure of heterologous protein complex loaded Thermotoga maritima encapsulin capsid | | Descriptor: | Maritimacin | | Authors: | Xiong, X, Sun, C, Vago, F.S, Klose, T, Zhu, J, Jiang, W. | | Deposit date: | 2020-09-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM Structure of Heterologous Protein Complex Loaded Thermotoga Maritima Encapsulin Capsid.
Biomolecules, 10, 2020
|
|
7TPB
 
 | | p120RasGAP SH3 domain in complex with DLC1 RhoGAP domain | | Descriptor: | Ras GTPase-activating protein 1, Rho GTPase-activating protein 7 | | Authors: | Stiegler, A.L, Boggon, T.J, Chau, J.E, Vish, K.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SH3 domain regulation of RhoGAP activity: Crosstalk between p120RasGAP and DLC1 RhoGAP.
Nat Commun, 13, 2022
|
|
9BUL
 
 | |
8K70
 
 | |
7LJP
 
 | | Structure of Thermotoga maritima SmpB | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2021-01-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Thermotoga maritima SmpB reveals its C-terminal tail domain in a helical conformation mimicking that of a ribosome-bound state
To Be Published
|
|
8CEE
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CDU
 
 | | Rnase R bound to a 30S degradation intermediate (main state) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CED
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CEC
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CDV
 
 | | Rnase R bound to a 30S degradation intermediate (state II) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.73 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
1DD5
 
 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA RIBOSOME RECYCLING FACTOR, RRF | | Descriptor: | ACETIC ACID, RIBOSOME RECYCLING FACTOR | | Authors: | Selmer, M, Al-Karadaghi, S, Hirokawa, G, Kaji, A, Liljas, A. | | Deposit date: | 1999-11-08 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic.
Science, 286, 1999
|
|
5V8S
 
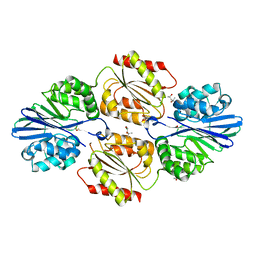 | | Flavo di-iron protein H90D mutant from Thermotoga maritima | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Taylor, A.B, Becker, A, Giri, N, Hart, P.J, Kurtz Jr, D.M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Flavo Di-iron protein H90D Mutant from Thermotoga Maritima
To Be Published
|
|
5XQO
 
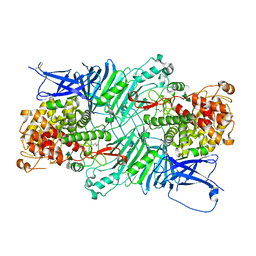 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with tetrameric substrate | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose-(1-4)-alpha-D-galactopyranuronic acid-(1-2)-alpha-L-rhamnopyranose, 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-alpha-L-rhamnopyranose-(1-4)-alpha-D-galactopyranuronic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, ... | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
7P92
 
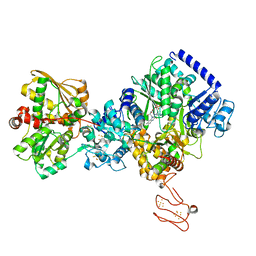 | | TmHydABC- T. maritima bifurcating hydrogenase with bridge domain up | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P8N
 
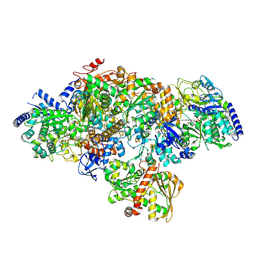 | | TmHydABC- T. maritima hydrogenase with bridge closed | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P91
 
 | | TmHydABC- T. maritima bifurcating hydrogenase with bridge domain closed | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P5H
 
 | | TmHydABC- D2 map | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-14 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
5XQG
 
 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, Pcrglx protein | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
6WKV
 
 | | Cryo-EM structure of engineered variant of the Encapsulin from Thermotoga maritima (TmE) | | Descriptor: | Encapsulin, FLAVIN MONONUCLEOTIDE | | Authors: | Williams, E, Jenkins, M, Zhao, H, Juneja, P, Lutz, S. | | Deposit date: | 2020-04-17 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of engineered variant of the Encapsulin from Thermotoga maritima (TmE)
To Be Published
|
|
8SUO
 
 | | BA.2/AZD1061/AZD3152 structure analysis | | Descriptor: | AZD1061 heavy chain, AZD1061 light chain, AZD3152 heavy chain, ... | | Authors: | Oganesyan, V, van Dyk, N, Dippel, A, Barnes, A, O'Connor, E. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray crystal structure of BA.2 RBD bound by two neutralizing antibodies
To Be Published
|
|
6PZ0
 
 | | Crystal structure of oxidized iodotyrosine deiodinase (IYD) bound to FMN and L-Tyrosine | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, TYROSINE, ... | | Authors: | Sun, Z, Kavran, J.M, Rokita, S.E. | | Deposit date: | 2019-07-31 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The minimal structure for iodotyrosine deiodinase function is defined by an outlier protein from the thermophilic bacterium Thermotoga neapolitana.
J.Biol.Chem., 297, 2021
|
|
6Q3U
 
 | | Gly52Ala mutant of arginine-bound ArgBP from T. maritima | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein, ... | | Authors: | Balasco, N, Smaldone, G, Ruggiero, A, Vitagliano, L. | | Deposit date: | 2018-12-04 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The characterization of Thermotoga maritima Arginine Binding Protein variants demonstrates that minimal local strains have an important impact on protein stability.
Sci Rep, 9, 2019
|
|
