5K70
 
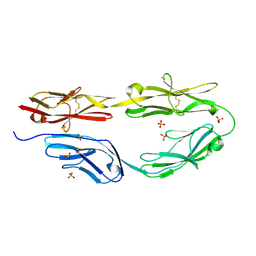 | | Sidekick-2 immunoglobulin domains 1-4 H18R/N22S mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
8Q81
 
 | | Photorhabdus laumondii lectin PLL3 in complex with alpha-methyl-fucoside | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Photorhabdus luminescens subsp. laumondii TTO1 complete genome segment 3/17, ... | | Authors: | Melicher, F, Houser, J, Paulenova, E, Wimmerova, M. | | Deposit date: | 2023-08-17 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The insight into the biology of five homologous lectins produced by the entomopathogenic bacterium and nematode symbiont Photorhabdus laumondii.
Glycobiology, 35, 2025
|
|
6VZ6
 
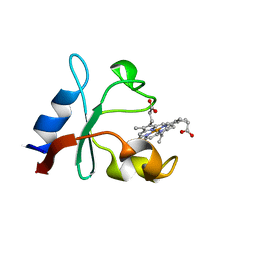 | | Methanococcoides burtonii cytochrome b5 domain protein (WP 011499504.1) | | Descriptor: | Cytochrome b5-domain protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Teakel, S.L, Forwood, J.K, Aragao, D, Cahill, M.A, Marama, M. | | Deposit date: | 2020-02-27 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methanococcoides burtonii cytochrome b5 domain protein (WP 011499504.1)
To Be Published
|
|
5NN1
 
 | |
5NJB
 
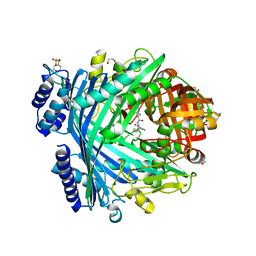 | | E. coli Microcin-processing metalloprotease TldD/E with actinonin bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACTINONIN, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
7SV4
 
 | | Crystal structure of SpaA-SLH in complex with 4,6-Pyr-beta-D-ManNAc-(1->4)-beta-D-GlcNAc-(1->3)-4,6-Pyr-beta-D-ManNAcOMe | | Descriptor: | GLYCEROL, Surface (S-) layer glycoprotein, methyl 2-acetamido-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl-(1->4)-2-acetamido-2-deoxy-beta-D-glucopyranosyl-(1->3)-2-acetamido-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranoside | | Authors: | Legg, M.S.G, Evans, S.V. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The S-layer homology domains of Paenibacillus alvei surface protein SpaA bind to cell wall polysaccharide through the terminal monosaccharide residue.
J.Biol.Chem., 298, 2022
|
|
9GCJ
 
 | |
6ZT6
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZG1
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
8EIG
 
 | | The complex of phosphorylated human delta F508 cystic fibrosis transmembrane conductance regulator (CFTR) with elexacaftor (VX-445) and ATP/Mg | | Descriptor: | (6P)-N-(1,3-dimethyl-1H-pyrazole-4-sulfonyl)-6-[3-(3,3,3-trifluoro-2,2-dimethylpropoxy)-1H-pyrazol-1-yl]-2-[(4S)-2,2,4-trimethylpyrrolidin-1-yl]pyridine-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, ... | | Authors: | Fiedorczuk, K, Chen, J. | | Deposit date: | 2022-09-15 | | Release date: | 2022-10-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular structures reveal synergistic rescue of Delta 508 CFTR by Trikafta modulators.
Science, 378, 2022
|
|
8EW3
 
 | | Cryo EM structure of Vibrio cholerae NQR | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit A, ... | | Authors: | Fuller, J.R, Juarez, O. | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.65159 Å) | | Cite: | Novel cofactor binding motifs, electron transfer and ion pumping mechanisms of the respiratory complex NQR
To Be Published
|
|
5IT8
 
 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-07-27 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9R59
 
 | | Crystal structure of MAPKAPK2 with a covalent compound GCL334 targeting lysine | | Descriptor: | 5-[3-[bis(oxidanyl)-$l^{3}-sulfanyl]oxy-4-chloranyl-phenyl]-~{N}-(4-piperazin-1-ylphenyl)-~{N}-(pyridin-2-ylmethyl)furan-2-carboxamide, MAP kinase-activated protein kinase 2 | | Authors: | Wang, G.Q, Hillebrand, L, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-05-08 | | Release date: | 2025-07-30 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A twist in the tale: shifting from covalent targeting of a tyrosine in JAK3 to a lysine in MK2.
Rsc Med Chem, 16, 2025
|
|
7Z5J
 
 | | The molybdenum storage protein loaded with tungstate | | Descriptor: | 1,1,3,3,5,7,7,9,11,15,15-undecakis($l^{1}-oxidanyl)-2$l^{4},4$l^{3},6$l^{5},8,10,12,14,16,17,18,19$l^{3},20,21,22,23-pentadecaoxa-1$l^{6},3$l^{6},5$l^{6},7$l^{6},9$l^{6},11$l^{6},13$l^{6},15$l^{6}-octatungstapentadecacyclo[7.7.1.1^{1,13}.1^{3,5}.1^{3,15}.1^{5,7}.1^{5,11}.1^{7,11}.0^{2,13}.0^{2,15}.0^{4,13}.0^{6,9}.0^{6,11}.0^{6,13}.0^{9,19}]tricosane, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ermler, U, Aziz, I, Kaltwasser, S, Kayastha, K, Vonck, J. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | The molybdenum storage protein forms and deposits distinct polynuclear tungsten oxygen aggregates.
J.Inorg.Biochem., 234, 2022
|
|
7NC9
 
 | | Glutathione-S-transferase GliG mutant H26N | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
9FOJ
 
 | | LGTV TP21. Langat virus, strain TP21 | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Bisikalo, K, Rosendal, E. | | Deposit date: | 2024-06-12 | | Release date: | 2024-11-06 | | Last modified: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Influence of the pre-membrane and envelope proteins on structure, pathogenicity, and tropism of tick-borne encephalitis virus.
J.Virol., 99, 2025
|
|
9FK0
 
 | | LGTV with TBEV prME | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Bisikalo, K, Rosendal, E. | | Deposit date: | 2024-06-01 | | Release date: | 2024-11-06 | | Last modified: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Influence of the pre-membrane and envelope proteins on structure, pathogenicity, and tropism of tick-borne encephalitis virus.
J.Virol., 99, 2025
|
|
7NCL
 
 | | Glutathione-S-transferase GliG mutant E82Q | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCD
 
 | | Glutathione-S-transferase GliG mutant N27D | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
9CFT
 
 | |
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
7NCO
 
 | | Glutathione-S-transferase GliG mutant K127R | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8C23
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200T (monoclinic form M200T#m) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
7NCP
 
 | | Glutathione-S-transferase GliG mutant K127A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG, SODIUM ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
