2NZ3
 
 | |
2NZ4
 
 | |
2NZ5
 
 | | Structure and Function Studies of Cytochrome P450 158A1 from Streptomyces coelicolor A3(2) | | Descriptor: | Cytochrome P450 CYP158A1, PROTOPORPHYRIN IX CONTAINING FE, naphthalene-1,2,4,5,7-pentol | | Authors: | Zhao, B, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2006-11-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Different binding modes of two flaviolin substrate molecules in cytochrome P450 158A1 (CYP158A1) compared to CYP158A2.
Biochemistry, 46, 2007
|
|
2NZ6
 
 | | Crystal structure of the PTPRJ inactivating mutant C1239S | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Ugochukwu, E, Barr, A, Savitsky, P, Pike, A.C.W, Bunkoczi, G, Sundstrom, M, Weigelt, J, Arrowsmith, C.H, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2NZ7
 
 | |
2NZ8
 
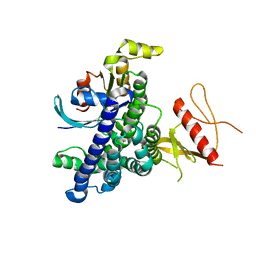 | | N-terminal DHPH cassette of Trio in complex with nucleotide-free Rac1 | | Descriptor: | ras-related C3 botulinum toxin substrate 1 isoform Rac1, triple functional domain protein | | Authors: | Chhatriwala, M.K, Betts, L, Worthylake, D.K, Sondek, J. | | Deposit date: | 2006-11-22 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The DH and PH Domains of Trio Coordinately Engage Rho GTPases for their Efficient Activation
J.Mol.Biol., 368, 2007
|
|
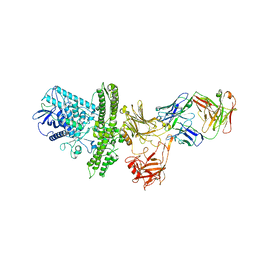
2NZ9
 
 | |
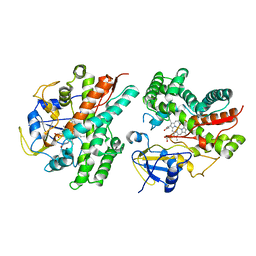
2NZA
 
 | |
2NZC
 
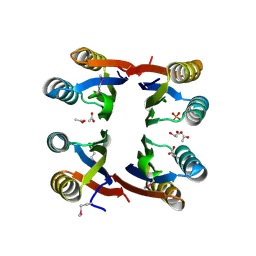 | | The structure of uncharacterized protein TM1266 from Thermotoga maritima. | | Descriptor: | ACETIC ACID, GLYCEROL, Hypothetical protein, ... | | Authors: | Cuff, M.E, Evdokimova, E, Kudritska, M, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of uncharacterized protein TM1266 from Thermotoga maritima.
TO BE PUBLISHED
|
|
2NZD
 
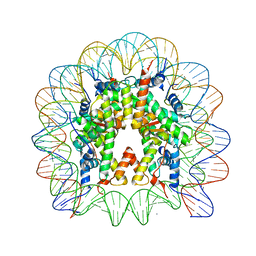 | | Nucleosome core particle containing 145 bp of DNA | | Descriptor: | DNA (145-MER), Histone H2B, Histone H3, ... | | Authors: | Ong, M.S, Richmond, T.J, Davey, C.A. | | Deposit date: | 2006-11-23 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA stretching and extreme kinking in the nucleosome core
J.Mol.Biol., 368, 2007
|
|
2NZE
 
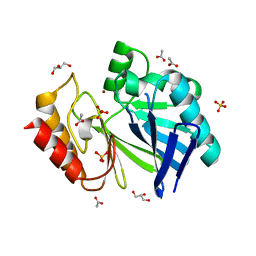 | | Structure of beta-lactamase II from Bacillus cereus. R121H, C221S double mutant. Space group P3121. | | Descriptor: | ACETIC ACID, Beta-lactamase II, GLYCEROL, ... | | Authors: | Medrano Martin, F.J, Vila, A.J, Gonzalez, J.M. | | Deposit date: | 2006-11-23 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Zn2 position in metallo-beta-lactamases is critical for activity: a study on chimeric metal sites on a conserved protein scaffold.
J.Mol.Biol., 373, 2007
|
|
2NZF
 
 | |
2NZH
 
 | |
2NZI
 
 | |
2NZJ
 
 | | The crystal structure of REM1 in complex with GDP | | Descriptor: | CHLORIDE ION, GTP-binding protein REM 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Turnbull, A.P, Papagrigoriou, E, Ugochukwu, E, Elkins, J.M, Soundararajan, M, Yang, X, Gorrec, F, Umeano, C, Salah, E, Burgess, N, Johansson, C, Berridge, G, Gileadi, O, Bray, J, Marsden, B, Watts, S, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C.H, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-23 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of REM1 in complex with GDP
To be Published
|
|
2NZL
 
 | | Crystal structure of human hydroxyacid oxidase 1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYOXYLIC ACID, Hydroxyacid oxidase 1 | | Authors: | Ugochukwu, E, Kavanagh, K, Pilka, E, Berridge, G, Debreczeni, J.E, Papagrigoriou, E, Turnbull, A, Niesen, F, Gileadi, O, von Delft, F, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-24 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of human hydroxyacid oxidase 1
To be Published
|
|
2NZM
 
 | | Hexasaccharide I bound to Bacillus subtilis pectate lyase | | Descriptor: | CALCIUM ION, Pectate lyase, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Pickersgill, R.W, Seyedarabi, A. | | Deposit date: | 2006-11-24 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into substrate specificity and the anti beta-elimination mechanism of pectate lyase.
Biochemistry, 49, 2010
|
|
2NZO
 
 | |
2NZT
 
 | | Crystal structure of human hexokinase II | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Hexokinase-2, UNKNOWN ATOM OR ION, ... | | Authors: | Rabeh, W.M, Zhu, H, Nedyalkova, L, Tempel, W, Wasney, G, Landry, R, Vedadi, M, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-25 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The catalytic inactivation of the N-half of human hexokinase 2 and structural and biochemical characterization of its mitochondrial conformation.
Biosci.Rep., 38, 2018
|
|
2NZU
 
 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | Authors: | Schumacher, M.A, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-11-25 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
2NZV
 
 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | Authors: | Schumacher, M.A, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-11-25 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
2NZW
 
 | | Crystal Structure of alpha1,3-Fucosyltransferase | | Descriptor: | Alpha1,3-fucosyltransferase, SULFATE ION | | Authors: | Sun, H.Y, Ko, T.P. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design.
J. Biol. Chem., 282, 2007
|
|
2NZX
 
 | | Crystal Structure of alpha1,3-Fucosyltransferase with GDP | | Descriptor: | Alpha1,3-fucosyltransferase, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Sun, H.Y, Ko, T.P. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design.
J. Biol. Chem., 282, 2007
|
|
2NZY
 
 | | Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose | | Descriptor: | Alpha1,3-Fucosyltransferase, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Sun, H.Y, Ko, T.P. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design.
J. Biol. Chem., 282, 2007
|
|
2NZZ
 
 | | NMR structure analysis of the Penetratin conjugated Gas (374-394) peptide | | Descriptor: | Penetratin conjugated Gas (374-394) peptide | | Authors: | D'Ursi, A.M, Rovero, P, Albrizio, S, Espsito, C, D'Errico, G, Novellino, E. | | Deposit date: | 2006-11-27 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Driving forces in the delivery of penetratin conjugated G protein fragment.
J.Med.Chem., 50, 2007
|
|
