1SDU
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | Descriptor: | ACETATE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, SULFATE ION, ... | | Authors: | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-02-14 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
1SG7
 
 | | NMR solution structure of the putative cation transport regulator ChaB | | Descriptor: | Putative Cation transport regulator chaB | | Authors: | Osborne, M.J, Siddiqui, N, Cygler, M, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-23 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ChaB, a putative membrane ion antiporter regulator from Escherichia coli
BMC STRUCT.BIOL., 4, 2004
|
|
2CWH
 
 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae complexed with NADPH and pyrrole-2-carboxylate | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRROLE-2-CARBOXYLATE, delta1-piperideine-2-carboxylate reductase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2005-06-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
1NAO
 
 | | SOLUTION STRUCTURE OF AN RNA 2'-O-METHYLATED RNA DUPLEX CONTAINING AN RNA/DNA HYBRID SEGMENT AT THE CENTER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA/RNA (5'-R(*OMGP*OMUP*OMC)-D(P*AP*TP*CP*T)-R(P*OMCP*OMC)-3'), RNA (5'-R(*GP*GP*AP*GP*AP*UP*GP*AP*C)-3') | | Authors: | Nishizaki, T, Iwai, S, Ohtsuka, E, Nakamura, H. | | Deposit date: | 1996-03-29 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA.2'-O-methylated RNA hybrid duplex containing an RNA.DNA hybrid segment at the center.
Biochemistry, 36, 1997
|
|
2CWI
 
 | | X-ray crystal structure analysis of recombinant wild-type canine milk lysozyme (apo-type) | | Descriptor: | Lysozyme C, milk isozyme, SULFATE ION | | Authors: | Akieda, D, Yasui, M, Aizawa, T, Yao, M, Watanabe, N, Tanaka, I, Demura, M, Kawano, K, Nitta, K. | | Deposit date: | 2005-06-20 | | Release date: | 2006-06-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Construction of an expression system of canine milk lysozyme in the methylotrophic yeast Pichia pastoris
To be Published
|
|
1NSO
 
 | | Folded monomer of protease from Mason-Pfizer monkey virus | | Descriptor: | Protease 13 kDa | | Authors: | Veverka, V, Bauerova, H, Zabransky, A, Lang, J, Ruml, T, Pichova, I, Hrabal, R. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a monomeric form of a retroviral protease
J.MOL.BIOL., 333, 2003
|
|
2CTR
 
 | | Solution structure of J-domain from human DnaJ subfamily B menber 9 | | Descriptor: | DnaJ homolog subfamily B member 9 | | Authors: | Kobayashi, N, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-24 | | Release date: | 2005-11-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of J-domain from human DnaJ subfamily B menber 9
To be Published
|
|
2CUA
 
 | | THE CUA DOMAIN OF CYTOCHROME BA3 FROM THERMUS THERMOPHILUS | | Descriptor: | DINUCLEAR COPPER ION, PROTEIN (CUA), ZINC ION | | Authors: | Williams, P.A, Blackburn, N.J, Sanders, D, Bellamy, H, Stura, E.A, Fee, J.A, Mcree, D.E. | | Deposit date: | 1999-02-18 | | Release date: | 1999-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The CuA domain of Thermus thermophilus ba3-type cytochrome c oxidase at 1.6 A resolution.
Nat.Struct.Biol., 6, 1999
|
|
1SJW
 
 | | Structure of polyketide cyclase SnoaL | | Descriptor: | METHYL 5,7-DIHYDROXY-2-METHYL-4,6,11-TRIOXO-3,4,6,11-TETRAHYDROTETRACENE-1-CARBOXYLATE, nogalonic acid methyl ester cyclase | | Authors: | Sultana, A, Kallio, P, Jansson, A, Wang, J.S, Neimi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation.
Embo J., 23, 2004
|
|
2CWF
 
 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, delta1-piperideine-2-carboxylate reductase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2005-06-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
1V32
 
 | | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g08430 from Arabidopsis thaliana | | Descriptor: | hypothetical protein RAFL09-47-K03 | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-24 | | Release date: | 2004-04-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g08430 from Arabidopsis thaliana
To be Published
|
|
2CWM
 
 | | Native Crystal Structure of NO releasing inductive lectin from seeds of the Canavalia maritima (ConM) | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, lectin | | Authors: | Cavada, B.S, De Azevedo Jr, W.F, Assreuy, A.M.S, Criddle, D.N, Gadelha, C.A.A, Delatorre, P, Souza, E.P, Rocha, B.A.M, Santi-Gadelha, T, Moreno, F.B.M.B. | | Deposit date: | 2005-06-22 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native crystal structure of a nitric oxide-releasing lectin from the seeds of Canavalia maritima
J.Struct.Biol., 152, 2005
|
|
1UUQ
 
 | | Exo-mannosidase from Cellvibrio mixtus | | Descriptor: | GLYCEROL, MANNOSYL-OLIGOSACCHARIDE GLUCOSIDASE, SULFATE ION | | Authors: | Dias, M.V.F, Vincent, F, Pell, G, Prates, J.A.M, Centeno, M.S.J, Ferreira, L.M.A, Gilbert, H.J, Davies, G.J, Fontes, C.M.G.A. | | Deposit date: | 2004-01-09 | | Release date: | 2004-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights Into the Molecular Determinants of Substrate Specificity in Glycoside Hydrolase Family 5 Revealed by the Crystal Structure and Kinetics of Cellvibrio Mixtus Mannosidase 5A
J.Biol.Chem., 279, 2004
|
|
1UP0
 
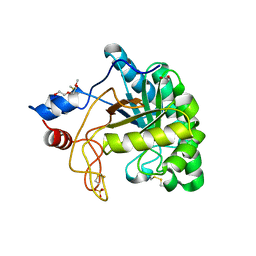 | | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with cellobiose at 1.75 angstrom | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, PUTATIVE CELLULASE CEL6, ... | | Authors: | Varrot, A, Leydier, S, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-09-26 | | Release date: | 2004-11-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mycobacterium Tuberculosis Strains Possess Functional Cellulases.
J.Biol.Chem., 280, 2005
|
|
2DK3
 
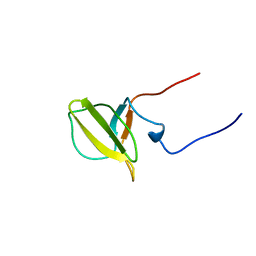 | | Solution structure of Mib-herc2 domain in HECT domain containing protein 1 | | Descriptor: | E3 ubiquitin-protein ligase HECTD1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Mib-herc2 domain in HECT domain containing protein 1
To be Published
|
|
1UWW
 
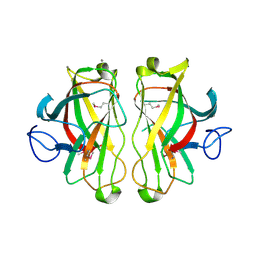 | | X-ray crystal structure of a non-crystalline cellulose specific carbohydrate-binding module: CBM28. | | Descriptor: | CALCIUM ION, ENDOGLUCANASE | | Authors: | Jamal, S, Nurizzo, D, Boraston, A, Davies, G.J. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-Ray Crystal Structure of a Non-Crystalline Cellulose-Specific Carbohydrate-Binding Module: Cbm28
J.Mol.Biol., 339, 2004
|
|
1Q8V
 
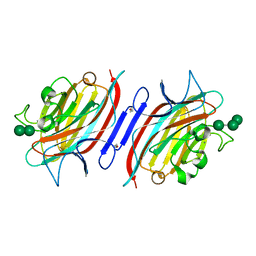 | | Pterocarpus angolensis lectin (PAL) in complex with the trimannoside [Man(Alpha1-3)]Man(alpha1-6)Man | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, ... | | Authors: | Loris, R, Van Walle, I, De Greve, H, Beeckmans, S, Deboeck, F, Wyns, L, Bouckaert, J. | | Deposit date: | 2003-08-22 | | Release date: | 2004-02-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Oligomannose Recognition by the Pterocarpus angolensis Seed Lectin
J.Mol.Biol., 335, 2004
|
|
1V6E
 
 | | Solution Structure of a N-terminal Ubiquitin-like Domain in Mouse Tubulin-specific Chaperone B | | Descriptor: | cytoskeleton-associated protein 1 | | Authors: | Zhao, C, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-29 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a N-terminal Ubiquitin-like Domain in Mouse Tubulin-specific Chaperone B
To be Published
|
|
1V1G
 
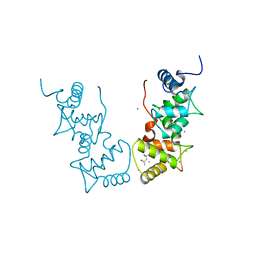 | | Structure of the Arabidopsis thaliana SOS3 complexed with Calcium(II) ion | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCINEURIN B-LIKE PROTEIN 4, CALCIUM ION, ... | | Authors: | Sanchez-Barrena, M.J, Martinez-Ripoll, M, Zhu, J.K, Albert, A. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of the Arabidopsis Thaliana SOS3: Molecular Mechanism of Sensing Calcium for Salt Stress Response
J.Mol.Biol., 345, 2005
|
|
1V3F
 
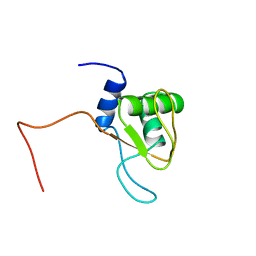 | |
1V4J
 
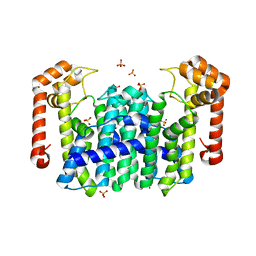 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima V73Y mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chou, C.C, Ko, T.P, Shr, H.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima and Mechanism of Product Chain Length Determination
J.Biol.Chem., 279, 2004
|
|
1V63
 
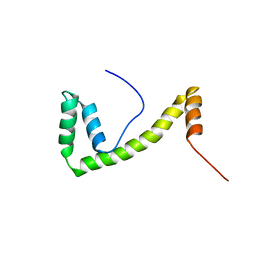 | | Solution structure of the 6th HMG box of mouse UBF1 | | Descriptor: | Nucleolar transcription factor 1 | | Authors: | Sato, M, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 6th HMG box of mouse UBF1
To be Published
|
|
1QGP
 
 | | NMR STRUCTURE OF THE Z-ALPHA DOMAIN OF ADAR1, 15 STRUCTURES | | Descriptor: | PROTEIN (DOUBLE STRANDED RNA ADENOSINE DEAMINASE) | | Authors: | Schade, M, Turner, C.J, Kuehne, R, Schmieder, P, Lowenhaupt, K, Herbert, A, Rich, A, Oschkinat, H. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QB4
 
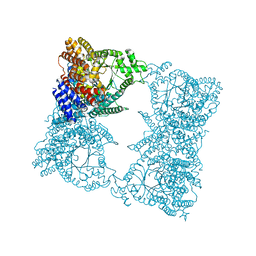 | | CRYSTAL STRUCTURE OF MN(2+)-BOUND PHOSPHOENOLPYRUVATE CARBOXYLASE | | Descriptor: | ASPARTIC ACID, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Matsumura, H, Terada, M, Shirakata, S, Inoue, T, Yoshinaga, T, Izui, K, Kai, Y. | | Deposit date: | 1999-04-30 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plausible phosphoenolpyruvate binding site revealed by 2.6 A structure of Mn2+-bound phosphoenolpyruvate carboxylase from Escherichia coli
FEBS Lett., 458, 1999
|
|
2DCQ
 
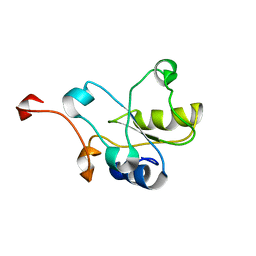 | |
