8ROZ
 
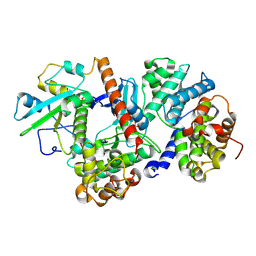 | |
8XU4
 
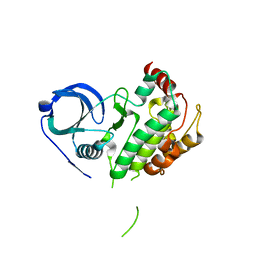 | | The Crystal Structure of MAPK2 from Biortus. | | Descriptor: | MALONIC ACID, MAP kinase-activated protein kinase 2 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Shen, Z. | | Deposit date: | 2024-01-12 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Crystal Structure of MAPK2 from Biortus.
To Be Published
|
|
8RO0
 
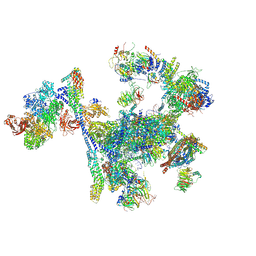 | | Structure of the C. elegans Intron Lariat Spliceosome primed for disassembly (ILS') | | Descriptor: | Cell division cycle 5-like protein, Coiled-coil domain-containing protein 12, GCF C-terminal domain-containing protein, ... | | Authors: | Vorlaender, M.K, Rothe, P, Plaschka, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
8RO1
 
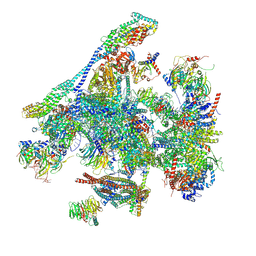 | | Structure of the C. elegans Intron Lariat Spliceosome double-primed for disassembly (ILS'') | | Descriptor: | CWF19-like protein 1 homolog, CWF19-like protein 2 homolog, Cell division cycle 5-like protein, ... | | Authors: | Vorlaender, M.K, Rothe, P, Plaschka, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
8RNW
 
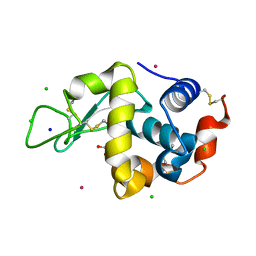 | | Hen Egg White Lysozyme soaked with trans-Ru(DMSO)4Cl2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
8VL0
 
 | |
8RNX
 
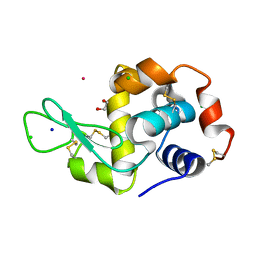 | | Hen Egg White Lysozyme soaked with [HIsq][trans-RuCl4(DMSO)(Isq)] | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
8RNV
 
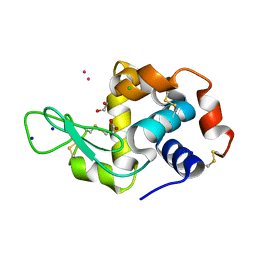 | | Hen Egg White Lysozyme soaked with cis-Ru(DMSO)4Cl2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
8RNY
 
 | | Hen Egg White Lysozyme soaked with with [H2Ind][trans-RuCl4(DMSO)(HInd)] | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
8XSX
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, E-tRNA, SERBP1 and eEF2 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8VKU
 
 | | Structure of VCP in complex with an ATPase activator (D2 domains only, hexameric form) | | Descriptor: | (3R)-N-[2-(ethylsulfanyl)phenyl]-3-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)butanamide, Transitional endoplasmic reticulum ATPase | | Authors: | Jones, N.H, Urnivicius, L, Kapoor, T.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Allosteric activation of VCP, an AAA unfoldase, by small molecule mimicry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VKF
 
 | |
8VKD
 
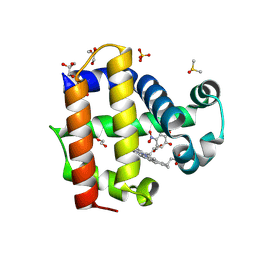 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VKC
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrophenol | | Descriptor: | Dehaloperoxidase A, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8XS3
 
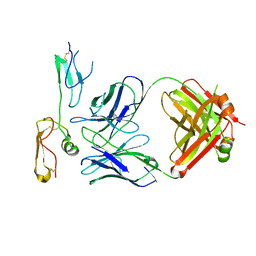 | | Structure of MPXV B6 and D68 fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D68_heavy chain, ... | | Authors: | wu, L.L, Sun, J.Q. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Two noncompeting human neutralizing antibodies targeting MPXV B6 show protective effects against orthopoxvirus infections.
Nat Commun, 15, 2024
|
|
8VK3
 
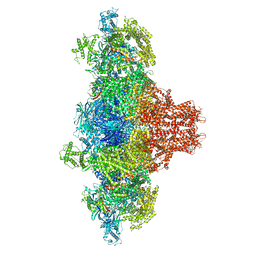 | |
8VK4
 
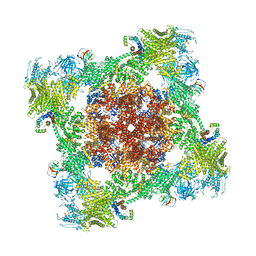 | |
8VJB
 
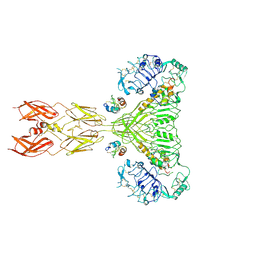 | | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8VJC
 
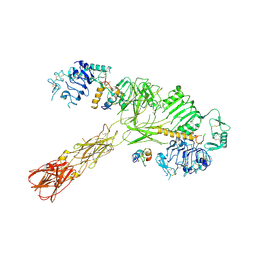 | | Cryo-EM structure of short form insulin receptor (IR-A) with three IGF2 bound, asymmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8VJ0
 
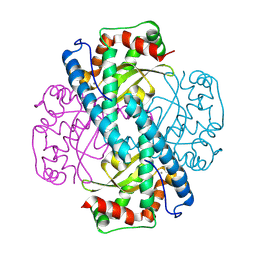 | | Neutron Structure of Oxidized Trp161Phe MnSOD | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-31 | | Method: | NEUTRON DIFFRACTION (2.3 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Biorxiv, 2024
|
|
8VJ4
 
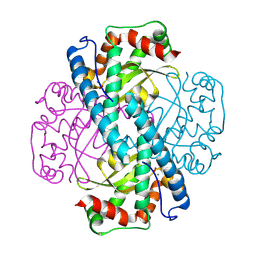 | | X-ray Counterpart to the Neutron Structure of Peroxide-Soaked Trp161Phe MnSOD | | Descriptor: | HYDROGEN PEROXIDE, MANGANESE (II) ION, Superoxide dismutase [Mn], ... | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Biorxiv, 2024
|
|
8VJ8
 
 | | X-ray Counterpart to Neutron Structure of Reduced Trp161Phe MnSOD | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Biorxiv, 2024
|
|
8VJ5
 
 | | Peroxide-Soaked MnSOD | | Descriptor: | HYDROGEN PEROXIDE, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Lutz, W.E, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Revealing the atomic and electronic mechanism of human manganese superoxide dismutase product inhibition.
Biorxiv, 2024
|
|
8VJ6
 
 | | GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 1 | | Descriptor: | 4-[(5S,8R)-8-methyl-6,7,8,9-tetrahydro-2H,5H-[1,3]dioxolo[4,5-h][2,3]benzodiazepin-5-yl]aniline, Isoform Flip of Glutamate receptor 2 | | Authors: | Hale, W.D, Montano Romero, A, Huganir, R.L, Twomey, E.C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Allosteric competition and inhibition in AMPA receptors.
Nat.Struct.Mol.Biol., 2024
|
|
8VJ7
 
 | | GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 2 | | Descriptor: | 4-[(5S,8R)-8-methyl-6,7,8,9-tetrahydro-2H,5H-[1,3]dioxolo[4,5-h][2,3]benzodiazepin-5-yl]aniline, GLUTAMIC ACID, Isoform Flip of Glutamate receptor 2 | | Authors: | Hale, W.D, Montano Romero, A, Huganir, R.L, Twomey, E.C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.85 Å) | | Cite: | Allosteric competition and inhibition in AMPA receptors.
Nat.Struct.Mol.Biol., 2024
|
|
