1WJZ
 
 | | Soluiotn structure of J-domain of mouse DnaJ like protein | | Descriptor: | 1700030A21Rik protein | | Authors: | Kobayashi, N, Koshiba, S, Inoue, M, Tochio, N, Tomizawa, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Soluiotn structure of J-domain of mouse DnaJ like protein
To be Published
|
|
1WLM
 
 | | Solution structure of mouse CGI-38 protein | | Descriptor: | Protein CGI-38 | | Authors: | Kobayashi, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-28 | | Release date: | 2005-07-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse CGI-38 protein
To be Published
|
|
2C36
 
 | | Structure of unliganded HSV gD reveals a mechanism for receptor- mediated activation of virus entry | | Descriptor: | CHLORIDE ION, GLYCOPROTEIN D HSV-1, ZINC ION, ... | | Authors: | Krummenacher, C, Supekar, V.M, Whitbeck, J.C, Lazear, E, Connolly, S.A, Eisenberg, R.J, Cohen, G.H, Wiley, D.C, Carfi, A. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry.
EMBO J., 24, 2005
|
|
1MUI
 
 | | Crystal structure of HIV-1 protease complexed with Lopinavir. | | Descriptor: | N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, protease | | Authors: | Stoll, V, Qin, W, Stewart, K.D, Jakob, C, Park, C, Walter, K, Simmer, R.L, Helfrich, R, Bussiere, D, Kao, J, Kempf, D, Sham, H.L, Norbeck, D.W. | | Deposit date: | 2002-09-23 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallographic Structure of ABT-378 (Lopinavir) Bound to HIV-1 Protease
BIOORG.MED.CHEM., 10, 2002
|
|
1WT6
 
 | | Coiled-Coil domain of DMPK | | Descriptor: | Myotonin-protein kinase | | Authors: | Garcia, P, Marino, M, Mayans, O. | | Deposit date: | 2004-11-16 | | Release date: | 2005-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into the self-assembly mechanism of dystrophia myotonica kinase
Faseb J., 20, 2006
|
|
1MVG
 
 | | NMR solution structure of chicken Liver basic Fatty Acid Binding Protein (Lb-FABP) | | Descriptor: | Liver basic Fatty Acid Binding Protein | | Authors: | Vasile, F, Ragona, L, Catalano, M, Zetta, L, Perduca, M, Monaco, H, Molinari, H. | | Deposit date: | 2002-09-25 | | Release date: | 2003-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of chicken Liver basic type Fatty Acid Binding Protein
J.BIOMOL.NMR, 25, 2003
|
|
1WXO
 
 | | Structure of Archaeal Trans-Editing Protein AlaX in complex with zinc | | Descriptor: | ZINC ION, alanyl-tRNA synthetase | | Authors: | Sokabe, M, Okada, A, Nakashima, T, Yao, M, Tanaka, I. | | Deposit date: | 2005-01-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular basis of alanine discrimination in editing site
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2BDD
 
 | | Crystal Structure of Holo-ACP-synthase from Plasmodium yoelii | | Descriptor: | ACP-synthase | | Authors: | Dong, A, Melone, M, Zhao, Y, Lew, J, Koeieradzki, I, Alam, Z, Wasney, G, Vedadi, M, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1MZ6
 
 | | Trypanosoma rangeli sialidase in complex with the inhibitor DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, sialidase | | Authors: | Buschiazzo, A, Tavares, G.A, Campetella, O, Spinelli, S, Cremona, M.L, Paris, G, Amaya, M.F, Frasch, A.C.C, Alzari, P.M. | | Deposit date: | 2002-10-05 | | Release date: | 2002-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of sialyltransferase activity in trypanosomal sialidases
Embo J., 19, 2000
|
|
1N27
 
 | | Solution structure of the PWWP domain of mouse Hepatoma-derived growth factor, related protein 3 | | Descriptor: | Hepatoma-derived growth factor, related protein 3 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-22 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PWWP domain of the hepatoma-derived growth factor family.
Protein Sci., 14, 2005
|
|
1N3Q
 
 | | Pterocarpus angolensis lectin complexed with turanose | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, alpha-D-glucopyranose-(1-3)-beta-D-fructofuranose, ... | | Authors: | Loris, R, Imberty, A, Beeckmans, S, Van Driessche, E, Read, J.S, Bouckaert, J, De Greve, H, Buts, L, Wyns, L. | | Deposit date: | 2002-10-29 | | Release date: | 2002-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Pterocarpus angolensis lectin in complex with glucose, sucrose, and turanose
J.BIOL.CHEM., 278, 2003
|
|
2BLQ
 
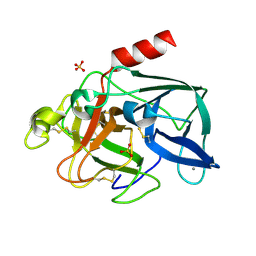 | | Elastase After A High Dose X-Ray "Burn" | | Descriptor: | CALCIUM ION, ELASTASE 1, SULFATE ION | | Authors: | Nanao, M.H, Ravelli, R.B. | | Deposit date: | 2005-03-08 | | Release date: | 2005-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Improving Radiation-Damage Substructures for Rip.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1X4G
 
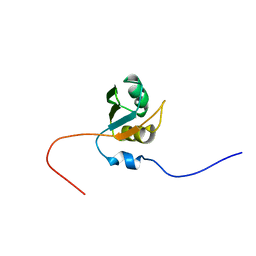 | | Solution structure of RRM domain in Nucleolysin TIAR | | Descriptor: | Nucleolysin TIAR | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in Nucleolysin TIAR
To be Published
|
|
1MUU
 
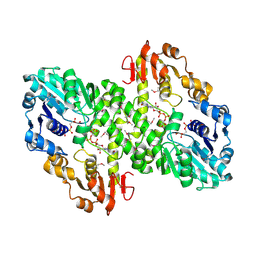 | | 2.0 A crystal structure of GDP-mannose dehydrogenase | | Descriptor: | GDP-mannose 6-dehydrogenase, GUANOSINE 5'-(TRIHYDROGEN DIPHOSPHATE), P'-D-MANNOPYRANOSYL ESTER, ... | | Authors: | Snook, C.F, Tipton, P.A, Beamer, L.J. | | Deposit date: | 2002-09-24 | | Release date: | 2003-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of GDP-mannose dehydrogenase: A key enzyme of alginate biosynthesis in P. aeruginosa
Biochemistry, 42, 2003
|
|
1MS3
 
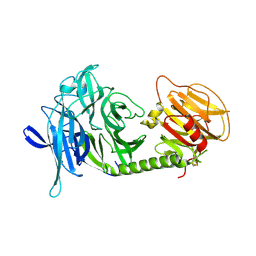 | | Monoclinic form of Trypanosoma cruzi trans-sialidase | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1X48
 
 | | Solution structure of the second DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | He, F, Muto, Y, Inoue, M, Tarada, T, Shirouzu, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase
To be Published
|
|
1X4O
 
 | | Solution structure of SURP domain in splicing factor 4 | | Descriptor: | Splicing factor 4 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SURP domain in splicing factor 4
To be Published
|
|
278D
 
 | |
1WE7
 
 | | Solution structure of Ubiquitin-like domain in SF3a120 | | Descriptor: | Sf3a1 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ubiquitin-like domain in SF3a120
To be Published
|
|
1WFR
 
 | | Solution structure of the conserved hypothetical protein TT1886, possibly sterol carrier protein, from Thermus Thermophilus HB8 | | Descriptor: | Hypothetical Protein TT1886 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the conserved hypothetical protein TT1886, possibly sterol carrier protein, from Thermus Thermophilus HB8
To be Published
|
|
1WI5
 
 | | Solution structure of the S1 RNA binding domain from human hypothetical protein BAA11502 | | Descriptor: | RRP5 protein homolog | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the S1 RNA binding domain from human hypothetical protein BAA11502
To be Published
|
|
1MS4
 
 | | Triclinic form of Trypanosoma cruzi trans-sialidase | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
220L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
225L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, PARA-XYLENE, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
232L
 
 | | T4 LYSOZYME MUTANT M120K | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Drew, D.L, Gassner, N, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
