1IIW
 
 | |
1WK1
 
 | | Solution structure of Lectin C-type domain derived from a hypothetical protein from C. elegans | | Descriptor: | Hypothetical protein yk1067a12 | | Authors: | Kobayashi, N, Koshiba, S, Inoue, M, Tochio, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Lectin C-type domain derived from a hypothetical protein from C. elegans
To be Published
|
|
1X4D
 
 | | Solution structure of RRM domain in Matrin 3 | | Descriptor: | Matrin 3 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in Matrin 3
To be Published
|
|
1IH8
 
 | | NH3-dependent NAD+ Synthetase from Bacillus subtilis Complexed with AMP-CPP and Mg2+ ions. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) synthetase | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, DeLucas, L. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1WWQ
 
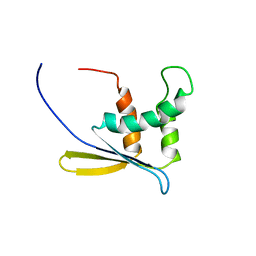 | | Solution Structure of Mouse ER | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mouse enhancer of rudimentary protein reveals a novel fold
J.Biomol.Nmr, 32, 2005
|
|
1WXS
 
 | | Solution Structure of Ufm1, a ubiquitin-fold modifier | | Descriptor: | Ubiquitin-fold Modifier 1 | | Authors: | Sasakawa, H, Sakata, E, Yamaguchi, Y, Komatsu, M, Tatsumi, K, Kominami, E, Tanaka, K, Kato, K. | | Deposit date: | 2005-02-01 | | Release date: | 2006-04-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Ufm1, a ubiquitin-fold modifier 1
Biochem.Biophys.Res.Commun., 343, 2006
|
|
1X4F
 
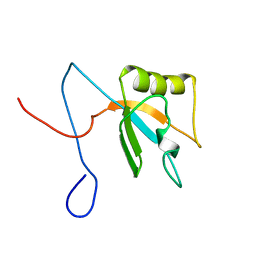 | | Solution structure of the second RRM domain in Matrin 3 | | Descriptor: | Matrin 3 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RRM domain in Matrin 3
To be Published
|
|
1IOQ
 
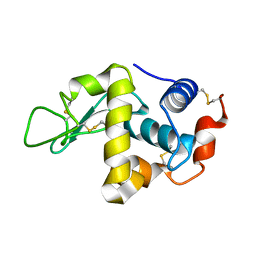 | | STABILIZATION OF HEN EGG WHITE LYSOZYME BY A CAVITY-FILLING MUTATION | | Descriptor: | LYSOZYME C | | Authors: | Ohmura, T, Ueda, T, Ootsuka, K, Saito, M, Imoto, T. | | Deposit date: | 2001-03-28 | | Release date: | 2001-04-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Stabilization of hen egg white lysozyme by a cavity-filling mutation.
Protein Sci., 10, 2001
|
|
1IOS
 
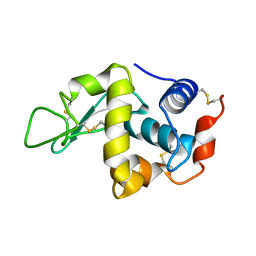 | | STABILIZATION OF HEN EGG WHITE LYSOZYME BY A CAVITY-FILLING MUTATION | | Descriptor: | LYSOZYME C | | Authors: | Ohmura, T, Ueda, T, Ootsuka, K, Saito, M, Imoto, T. | | Deposit date: | 2001-03-28 | | Release date: | 2001-04-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Stabilization of hen egg white lysozyme by a cavity-filling mutation.
Protein Sci., 10, 2001
|
|
1J2U
 
 | | Creatininase Zn | | Descriptor: | SULFATE ION, ZINC ION, creatinine amidohydrolase | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1J7T
 
 | |
1WFQ
 
 | | Solution structure of the first cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
1WH2
 
 | | Solution structure of the GYF domain of a hypothetical protein from Arabidopsis thaliana | | Descriptor: | Hypothetical protein At5g08430 | | Authors: | Nameki, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the GYF domain of a hypothetical protein from Arabidopsis thaliana
To be Published
|
|
1IGU
 
 | |
1IJ0
 
 | |
1WJZ
 
 | | Soluiotn structure of J-domain of mouse DnaJ like protein | | Descriptor: | 1700030A21Rik protein | | Authors: | Kobayashi, N, Koshiba, S, Inoue, M, Tochio, N, Tomizawa, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Soluiotn structure of J-domain of mouse DnaJ like protein
To be Published
|
|
1X48
 
 | | Solution structure of the second DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | He, F, Muto, Y, Inoue, M, Tarada, T, Shirouzu, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase
To be Published
|
|
1WLM
 
 | | Solution structure of mouse CGI-38 protein | | Descriptor: | Protein CGI-38 | | Authors: | Kobayashi, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-28 | | Release date: | 2005-07-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse CGI-38 protein
To be Published
|
|
1X4O
 
 | | Solution structure of SURP domain in splicing factor 4 | | Descriptor: | Splicing factor 4 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SURP domain in splicing factor 4
To be Published
|
|
1IN3
 
 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1X4G
 
 | | Solution structure of RRM domain in Nucleolysin TIAR | | Descriptor: | Nucleolysin TIAR | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in Nucleolysin TIAR
To be Published
|
|
1WE7
 
 | | Solution structure of Ubiquitin-like domain in SF3a120 | | Descriptor: | Sf3a1 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ubiquitin-like domain in SF3a120
To be Published
|
|
1WFR
 
 | | Solution structure of the conserved hypothetical protein TT1886, possibly sterol carrier protein, from Thermus Thermophilus HB8 | | Descriptor: | Hypothetical Protein TT1886 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the conserved hypothetical protein TT1886, possibly sterol carrier protein, from Thermus Thermophilus HB8
To be Published
|
|
1IOR
 
 | | STABILIZATION OF HEN EGG WHITE LYSOZYME BY A CAVITY-FILLING MUTATION | | Descriptor: | LYSOZYME C | | Authors: | Ohmura, T, Ueda, T, Ootsuka, K, Saito, M, Imoto, T. | | Deposit date: | 2001-03-28 | | Release date: | 2001-04-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Stabilization of hen egg white lysozyme by a cavity-filling mutation.
Protein Sci., 10, 2001
|
|
1WI5
 
 | | Solution structure of the S1 RNA binding domain from human hypothetical protein BAA11502 | | Descriptor: | RRP5 protein homolog | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the S1 RNA binding domain from human hypothetical protein BAA11502
To be Published
|
|
