3GW4
 
 | | Crystal structure of uncharacterized protein from Deinococcus radiodurans. Northeast Structural Genomics Consortium Target DrR162B. | | Descriptor: | Uncharacterized protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Northeast Structural Genomics Consortium Target DrR162B
To be published
|
|
2V7G
 
 | |
3DE1
 
 | |
3H5U
 
 | | Hepatitis C virus polymerase NS5B with saccharin inhibitor 1 | | Descriptor: | N-({3-[(5S)-5-tert-butyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-1,2-benzisothiazol-7-yl}methyl)methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 4: structure-based design, synthesis, and biological evaluation of benzo[d]isothiazole-1,1-dioxides
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2V50
 
 | |
3DDZ
 
 | |
2V2V
 
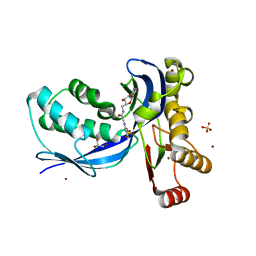 | | IspE in complex with ligand | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, 5'-[(1H-BENZIMIDAZOL-2-YLACETYL)AMINO]-5'-DEOXYCYTIDINE, BROMIDE ION, ... | | Authors: | Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-06-07 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and Characterization of Cytidine Derivatives that Inhibit the Kinase Ispe of the Non-Mevalonate Pathway for Isoprenoid Biosynthesis.
Chemmedchem, 3, 2008
|
|
3H59
 
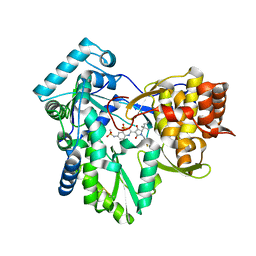 | | Hepatitis C virus polymerase NS5B with thiazine inhibitor 2 | | Descriptor: | N-{3-[(5S)-5-(1,1-dimethylpropyl)-1-(4-fluoro-3-methylbenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-4H-1,4-benzothiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-04-21 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 3: synthesis and optimization studies of benzothiazine-substituted tetramic acids
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3D9Q
 
 | |
3DE0
 
 | |
3HCI
 
 | | Structure of MsrB from Xanthomonas campestris (complex-like form) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, CALCIUM ION, Peptide methionine sulfoxide reductase, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
3BQH
 
 | |
3H8E
 
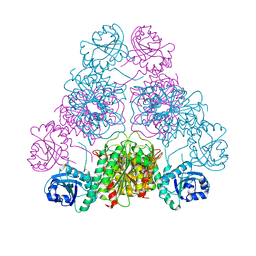 | |
3I6H
 
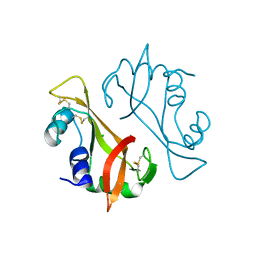 | | Ribonuclease A by LB nanotemplate method before high X-Ray dose on ESRF ID14-2 beamline | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic structure and radiation resistance of langmuir-blodgett protein crystals
To be Published
|
|
3BUK
 
 | | Crystal Structure of the Neurotrophin-3 and p75NTR Symmetrical Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neurotrophin-3, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Jiang, T, Gong, Y, Cao, P, Yu, H.J. | | Deposit date: | 2008-01-02 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the neurotrophin-3 and p75NTR symmetrical complex.
Nature, 454, 2008
|
|
2PNJ
 
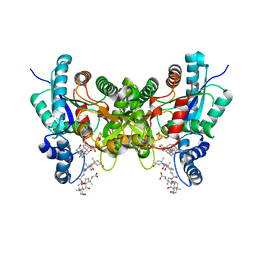 | | Crystal structure of human ferrochelatase mutant with Phe 337 replaced by Ala | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | Authors: | Dailey, H.A, Wu, C.-K, Horanyi, P, Medlock, A.E, Najahi-Missaoui, W, Burden, A.E, Dailey, T.A, Rose, J.P. | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Altered orientation of active site residues in variants of human ferrochelatase. Evidence for a hydrogen bond network involved in catalysis
Biochemistry, 46, 2007
|
|
3I5B
 
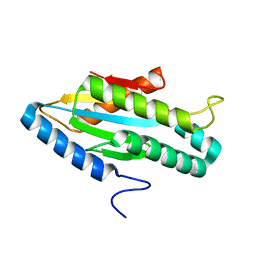 | |
2PXW
 
 | | Crystal Structure of N66D Mutant of Green Fluorescent Protein from Zoanthus sp. at 2.4 A Resolution (Transition State) | | Descriptor: | GFP-like fluorescent chromoprotein FP506 | | Authors: | Pletnev, S.V, Pletneva, N.V, Tikhonova, T.V, Pletnev, V.Z. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3C2H
 
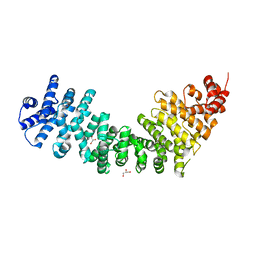 | | Crystal Structure of SYS-1 at 2.6A resolution | | Descriptor: | CITRATE ANION, GLYCEROL, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
3I6C
 
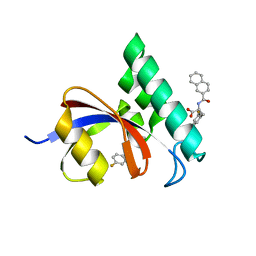 | | Structure-Based Design of Novel PIN1 Inhibitors (II) | | Descriptor: | 3-fluoro-N-(naphthalen-2-ylcarbonyl)-D-phenylalanine, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Greasley, S.E, Ferre, R.A. | | Deposit date: | 2009-07-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (II).
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3C7W
 
 | |
3HWU
 
 | |
2Q8H
 
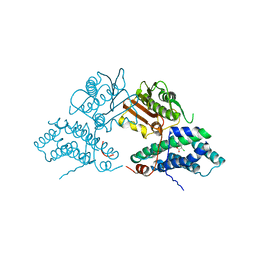 | | Structure of pyruvate dehydrogenase kinase isoform 1 in complex with dichloroacetate (DCA) | | Descriptor: | DICHLORO-ACETIC ACID, POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
2Q0R
 
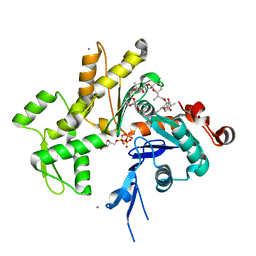 | | Structure of Pectenotoxin-2 Bound to Actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, CALCIUM ION, ... | | Authors: | Allingham, J.S, Miles, C.O, Rayment, I. | | Deposit date: | 2007-05-22 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural basis for regulation of actin polymerization by pectenotoxins.
J.Mol.Biol., 371, 2007
|
|
3BX7
 
 | |
