2SGA
 
 | |
3FZ9
 
 | | Crystal structure of poplar glutaredoxin S12 in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutaredoxin | | Authors: | Didierjean, C, Corbier, C, Koh, C.S, Rouhier, N, Jacquot, J.P. | | Deposit date: | 2009-01-24 | | Release date: | 2009-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site.
J.Biol.Chem., 284, 2009
|
|
3G0J
 
 | | Crystal Structure of the fifth Bromodomain of Human Poly-bromodomain containing protein 1 (PB1) | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Keates, T, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-28 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the fifth Bromodomain of Human Poly-bromodomain containing protein 1 (PB1)
To be Published
|
|
3GFX
 
 | | Klebsiella pneumoniae BlrP1 pH 4.5 calcium/cy-diGMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Barends, T, Hartmann, E, Griese, J, Beitlich, T, Kirienko, N, Ryjenkov, D, Reinstein, J, Shoeman, R, Gomelsky, M, Schlichting, I. | | Deposit date: | 2009-02-27 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase.
Nature, 459, 2009
|
|
3G1J
 
 | | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS4. | | Descriptor: | Integron cassette protein | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Tan, K, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS4.
To be published
|
|
3CFS
 
 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | Descriptor: | ARSENIC, Histone H4, Histone-binding protein RBBP7 | | Authors: | Murzina, N.V, Pei, X.-Y, Pratap, J.V, Sparkes, M, Vicente-Garcia, J, Ben-Shahar, T.R, Verreault, A, Luisi, B.F, Laue, E.D. | | Deposit date: | 2008-03-04 | | Release date: | 2008-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
2R3F
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 5-(2,3-dichlorophenyl)-N-(pyridin-4-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3Q
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-((3-bromo-5-o-tolylpyrazolo[1,5-a]pyrimidin-7-ylamino)methyl)pyridine 1-oxide, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
3G6R
 
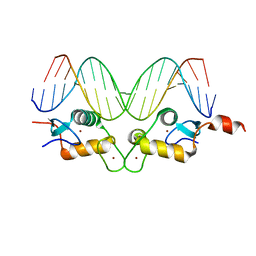 | | GR DNA binding domain:FKBP5 complex-52, 18bp | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*CP*CP*CP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*GP*GP*TP*GP*TP*TP*CP*TP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-08 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
2R4T
 
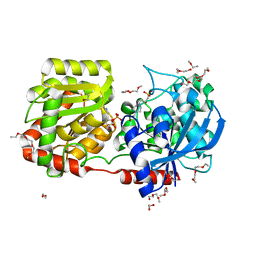 | | Crystal Structure of Wild-type E.coli GS in Complex with ADP and Glucose(wtGSc) | | Descriptor: | (2R)-2-hydroxy-3-[4-(2-hydroxyethyl)piperazin-1-yl]propane-1-sulfonic acid, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sheng, F, Geiger, J. | | Deposit date: | 2007-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.258 Å) | | Cite: | The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase.
J.Biol.Chem., 284, 2009
|
|
2QZQ
 
 | | Crystal structure of C-terminal of Aida | | Descriptor: | Axin interactor, dorsalization associated protein | | Authors: | Zheng, L.S, Wu, J.W. | | Deposit date: | 2007-08-17 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of C-terminal of Aida
To be Published
|
|
3GBZ
 
 | |
3CKK
 
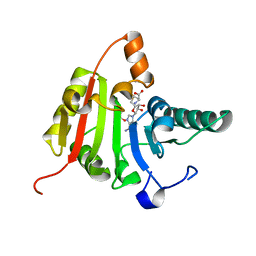 | | Crystal structure of human methyltransferase-like protein 1 | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Dong, A, Zeng, H, Dobrovetsky, E, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-16 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of human methyltransferase-like protein 1.
To be Published
|
|
2R3I
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 5-(2-fluorophenyl)-N-(pyridin-4-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
3GG1
 
 | | Klebsiella pneumoniae BlrP1 pH 8.0 calcium/cy-diGMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Barends, T, Hartmann, E, Griese, J, Beitlich, T, Kirienko, N, Ryjenkov, D, Reinstein, J, Shoeman, R, Gomelsky, M, Schlichting, I. | | Deposit date: | 2009-02-27 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase.
Nature, 459, 2009
|
|
3CXQ
 
 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
2TOB
 
 | | SOLUTION STRUCTURE OF THE TOBRAMYCIN-RNA APTAMER COMPLEX, NMR, 13 STRUCTURES | | Descriptor: | 1,3-DIAMINO-4,5,6-TRIHYDROXY-CYCLOHEXANE, 2,6-diammonio-2,3,6-trideoxy-alpha-D-glucopyranose, 3-ammonio-3-deoxy-alpha-D-glucopyranose, ... | | Authors: | Jiang, L, Patel, D.J. | | Deposit date: | 1998-06-17 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tobramycin-RNA aptamer complex.
Nat.Struct.Biol., 5, 1998
|
|
3G9K
 
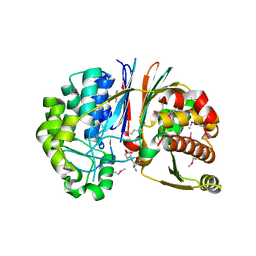 | | Crystal structure of Bacillus anthracis transpeptidase enzyme CapD | | Descriptor: | Capsule biosynthesis protein capD, GLUTAMIC ACID | | Authors: | Zhang, R, Wu, R, Richter, S, Anderson, V.J, Missiakas, D, Joachimiak, A. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of Bacillus anthracis Transpeptidase Enzyme CapD.
J.Biol.Chem., 284, 2009
|
|
3GFW
 
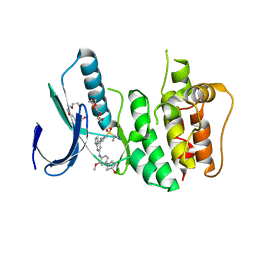 | | Crystal Structure of Human Dual Specificity Protein Kinase (TTK) in complex with a pyrolo-pyridin ligand | | Descriptor: | 1-(4-(4-(2-(isopropylsulfonyl)phenylamino)-1H-pyrrolo[2,3-b]pyridin-6-ylamino)-3-methoxyphenyl)piperidin-4-ol, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Filippakopoulos, P, Soundararajan, M, Choi, H, Keates, T, Elkins, J.M, King, O, Fedorov, O, Picaud, S.S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Grey, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function.
Nat.Chem.Biol., 6, 2010
|
|
3GH5
 
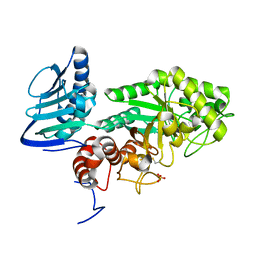 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, beta-hexosaminidase | | Authors: | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-03 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
3GG0
 
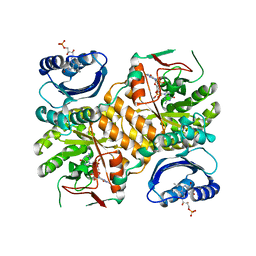 | | Klebsiella pneumoniae BlrP1 pH 9.0 manganese/cy-diGMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FLAVIN MONONUCLEOTIDE, Klebsiella pneumoniae BlrP1, ... | | Authors: | Barends, T, Hartmann, E, Griese, J, Beitlich, T, Kirienko, N, Ryjenkov, D, Reinstein, J, Shoeman, R, Gomelsky, M, Schlichting, I. | | Deposit date: | 2009-02-27 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase.
Nature, 459, 2009
|
|
3BUS
 
 | | Crystal Structure of RebM | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCoy, J.G, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and mechanism of the rebeccamycin sugar 4'-O-methyltransferase RebM.
J.Biol.Chem., 283, 2008
|
|
3GH7
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, SULFATE ION, beta-hexosaminidase | | Authors: | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
2R6F
 
 | | Crystal Structure of Bacillus stearothermophilus UvrA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Excinuclease ABC subunit A, ZINC ION | | Authors: | Inuzuka, Y, Pakotiprapha, D, Bowman, B.R, Jeruzalmi, D, Verdine, G.L. | | Deposit date: | 2007-09-05 | | Release date: | 2008-01-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Bacillus stearothermophilus UvrA Provides Insight into ATP-Modulated Dimerization, UvrB Interaction, and DNA Binding.
Mol.Cell, 29, 2008
|
|
3BWT
 
 | |
