8ER5
 
 | | Crystal Structure of NlpC/P60 domain from Clostridium innocuum NlpC/P60 domain-containing protein CI_01448. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NlpC/P60 domain-containing protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of NlpC/P60 domain from Clostridium innocuum NlpC/P60 domain-containing protein CI_01448.
To Be Published
|
|
1EYP
 
 | | CHALCONE ISOMERASE | | Descriptor: | CHALCONE-FLAVONONE ISOMERASE 1 | | Authors: | Jez, J.M, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-05-08 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase.
Nat.Struct.Biol., 7, 2000
|
|
7NT4
 
 | | X-ray structure of SCoV2-PLpro in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, PROFLAVIN, ... | | Authors: | Napolitano, V, Mourao, A, Bostock, M, Matsuda, A, Czarna, A, Popowicz, G.M. | | Deposit date: | 2021-03-09 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Acriflavine, a clinically approved drug, inhibits SARS-CoV-2 and other betacoronaviruses.
Cell Chem Biol, 29, 2022
|
|
7O04
 
 | |
1ECN
 
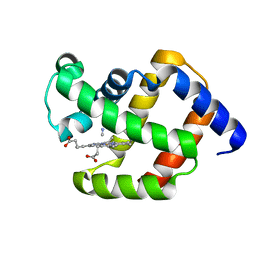 | |
1E65
 
 | |
1E4S
 
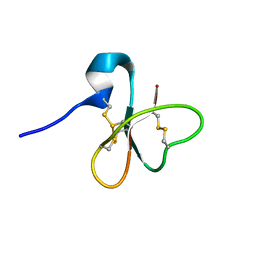 | | Solution structure of the human defensin hBD-1 | | Descriptor: | BETA-DEFENSIN 1 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of Human and Murine Beta-Defensins Reveals Structural Conservation in the Absence of Significant Sequence Similarity
Protein Sci., 10, 2001
|
|
8OQX
 
 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK with a phosphate analogue | | Descriptor: | 1,2-ETHANEDIOL, ATPase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
303D
 
 | | META-HYDROXY ANALOGUE OF HOECHST 33258 ('HYDROXYL OUT' CONFORMATION) BOUND TO D(CGCGAATTCGCG)2 | | Descriptor: | 3-[5-[5-(4-METHYL-PIPERAZIN-1-YL)-1H-IMIDAZO[4,5-B]PYRIDIN-2-YL]-BENZIMIDAZOL-2-YL]-PHENOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Clark, G.R, Squire, C.J, Gray, E.J, Leupin, W, Neidle, S. | | Deposit date: | 1996-06-26 | | Release date: | 1997-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Designer DNA-binding drugs: the crystal structure of a meta-hydroxy analogue of Hoechst 33258 bound to d(CGCGAATTCGCG)2.
Nucleic Acids Res., 24, 1996
|
|
1F23
 
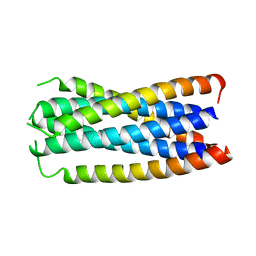 | | CONTRIBUTION OF A BURIED HYDROGEN BOND TO HIV-1 ENVELOPE GLYCOPROTEIN STRUCTURE AND FUNCTION | | Descriptor: | TRANSMEMBRANE GLYCOPROTEIN | | Authors: | Liu, J, Shu, W, Fagan, M, Nunberg, J.H, Lu, M. | | Deposit date: | 2000-05-23 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of the HIV gp41 core containing an Ile573 to Thr substitution: implications for membrane fusion.
Biochemistry, 40, 2001
|
|
7O9Y
 
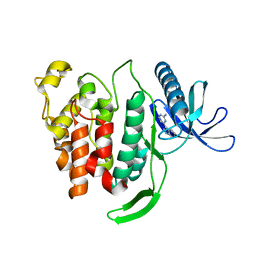 | |
1F0T
 
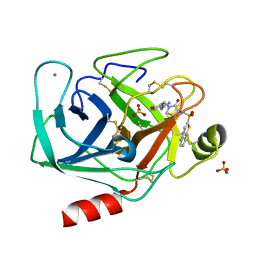 | | BOVINE TRYPSIN COMPLEXED WITH RPR131247 | | Descriptor: | 4-HYDROXY-3-[2-OXO-3-(THIENO[3,2-B]PYRIDINE-2-SULFONYLAMINO)-PYRROLIDIN-1-YLMETHYL]-BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Maignan, S, Guilloteau, J.P, Pouzieux, S, Choi-Sledeski, Y.M, Becker, M.R, Klein, S.I, Ewing, W.R, Pauls, H.W, Spada, A.P, Mikol, V. | | Deposit date: | 2000-05-17 | | Release date: | 2000-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of human factor Xa complexed with potent inhibitors.
J.Med.Chem., 43, 2000
|
|
1F0S
 
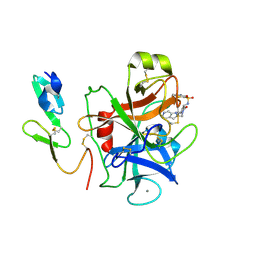 | | Crystal Structure of Human Coagulation Factor XA Complexed with RPR208707 | | Descriptor: | CALCIUM ION, COAGULATION FACTOR XA, THIENO[3,2-B]PYRIDINE-2-SULFONIC ACID [2-OXO-1-(1H-PYRROLO[2,3-C]PYRIDIN-2-YLMETHYL)-PYRROLIDIN-3-YL]-AMIDE | | Authors: | Maignan, S, Guilloteau, J.P, Pouzieux, S, Choi-Sledeski, Y.M, Becker, M.R, Klein, S.I, Ewing, W.R, Pauls, H.W, Spada, A.P, Mikol, V. | | Deposit date: | 2000-05-17 | | Release date: | 2000-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human factor Xa complexed with potent inhibitors.
J.Med.Chem., 43, 2000
|
|
8G0T
 
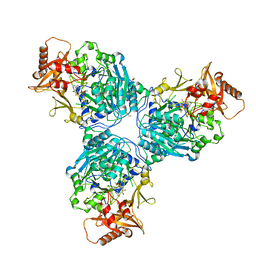 | |
7O9T
 
 | |
6N36
 
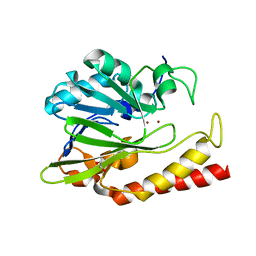 | | Beta-lactamase from Chitinophaga pinensis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, ZINC ION | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Beta-lactamase from Chitinophaga pinensis
to be published
|
|
7OAS
 
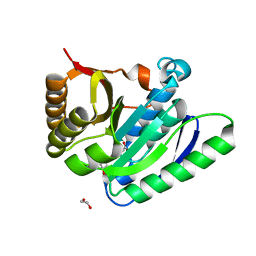 | | Structural basis for targeted p97 remodeling by ASPL as prerequisite for p97 trimethylation by METTL21D | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | | Authors: | Petrovic, S, Heinemann, U, Roske, Y. | | Deposit date: | 2021-04-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Structural remodeling of AAA+ ATPase p97 by adaptor protein ASPL facilitates posttranslational methylation by METTL21D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FEY
 
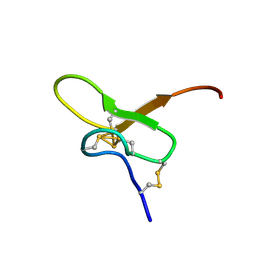 | | Solution structure of Pmu1a | | Descriptor: | Pmu1a toxin | | Authors: | Daly, N.L, Wilson, D.T. | | Deposit date: | 2022-12-07 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Pmu1a, a novel spider toxin with dual inhibitory activity at pain targets hNa V 1.7 and hCa V 3 voltage-gated channels.
Febs J., 290, 2023
|
|
8OKH
 
 | |
1F5W
 
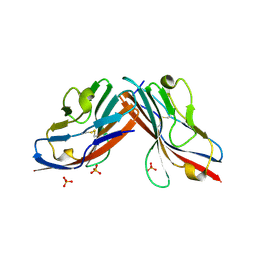 | | DIMERIC STRUCTURE OF THE COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR D1 DOMAIN | | Descriptor: | COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR, SULFATE ION | | Authors: | van Raaij, M.J, Chouin, E, van der Zandt, H, Bergelson, J.M, Cusack, S. | | Deposit date: | 2000-06-18 | | Release date: | 2000-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 A resolution.
Structure Fold.Des., 8, 2000
|
|
8P52
 
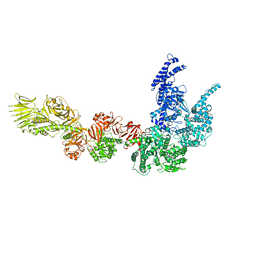 | |
7P36
 
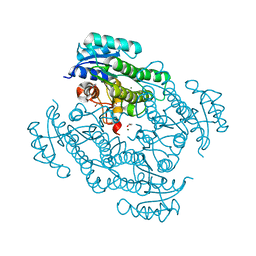 | | X-ray structure of Lactobacillus kefir alcohol dehydrogenase (wild type) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Transfer of a Rational Crystal Contact Engineering Strategy between Diverse Alcohol Dehydrogenases
Crystals, 11, 2021
|
|
1F6B
 
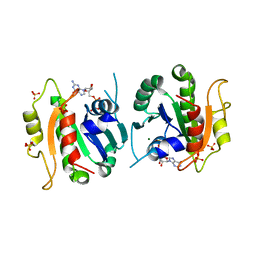 | | CRYSTAL STRUCTURE OF SAR1-GDP COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SAR1, ... | | Authors: | Huang, M, Wilson, I.A, Balch, W.E. | | Deposit date: | 2000-06-21 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH2 terminus in ER export.
J.Cell Biol., 155, 2001
|
|
7QDI
 
 | | Structure of octameric left-handed 310-helix bundle: D-310HD | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2021-11-27 | | Release date: | 2022-04-27 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
7P7Y
 
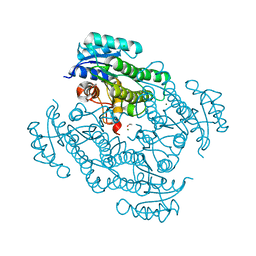 | | X-ray structure of Lactobacillus kefir alcohol dehydrogenase mutant Q126K | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Transfer of a Rational Crystal Contact Engineering Strategy between Diverse Alcohol Dehydrogenases
Crystals, 11, 2021
|
|
