4BEB
 
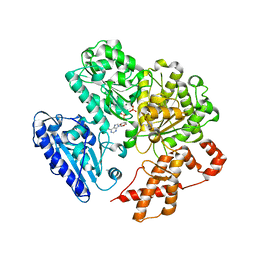 | | MUTANT (K220E) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
4B9P
 
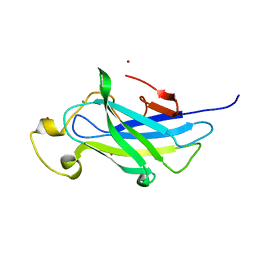 | | Biomass sensoring module from putative Rsgi2 protein of Clostridium thermocellum resemble family 3 carbohydrate-binding module of cellulosome | | Descriptor: | CALCIUM ION, TYPE 3A CELLULOSE-BINDING DOMAIN PROTEIN, ZINC ION | | Authors: | Yaniv, O, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.182 Å) | | Cite: | Fine-Structural Variance of Family 3 Carbohydrate-Binding Modules as Extracellular Biomass-Sensing Components of Clostridium Thermocellum Anti-Sigma(I) Factors.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3THH
 
 | |
1ULO
 
 | |
2GTG
 
 | |
4Q0J
 
 | | Deinococcus radiodurans BphP photosensory module | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Vierstra, R.D. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-16 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (2.747 Å) | | Cite: | Crystallographic and Electron Microscopic Analyses of a Bacterial Phytochrome Reveal Local and Global Rearrangements during Photoconversion.
J.Biol.Chem., 289, 2014
|
|
1Q3T
 
 | | Solution structure and function of an essential CMP kinase of Streptococcus pneumoniae | | Descriptor: | Cytidylate kinase | | Authors: | Yu, L, Mack, J, Hajduk, P.J, Kakavas, S.J, Saiki, A.Y, Lerner, C.G, Olejniczak, E.T. | | Deposit date: | 2003-07-31 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and function of an essential CMP kinase of Streptococcus pneumoniae
Protein Sci., 12, 2003
|
|
2L6Q
 
 | |
2HQF
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
2HQC
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
2HUA
 
 | |
2HI7
 
 | | Crystal structure of DsbA-DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Murakami, S, Suzuki, M, Nakagawa, A, Yamashita, E, Okada, K, Ito, K. | | Deposit date: | 2006-06-29 | | Release date: | 2006-12-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation
Cell(Cambridge,Mass.), 127, 2006
|
|
3PBY
 
 | | Crystal structure of the mutant L123S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
2LD2
 
 | |
3PBW
 
 | | Crystal structure of the mutant L123N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3TNW
 
 | | Structure of CDK2/cyclin A in complex with CAN508 | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Baumli, S, Hole, A.J, Endicott, J.A. | | Deposit date: | 2011-09-02 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
4F6H
 
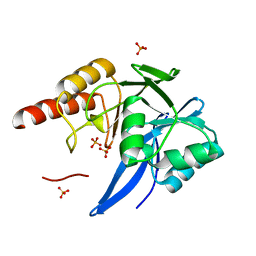 | | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-b-lactamase active site | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Horton, L.B, Shanker, S, Sankaran, B, Mikulski, R, Brown, N.G, Phillips, K, Lykissa, E, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2012-05-14 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site
Antimicrob.Agents Chemother., 56, 2012
|
|
3PBU
 
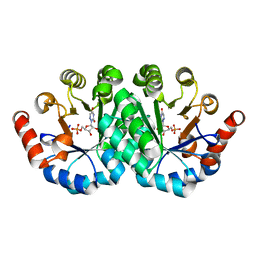 | | Crystal structure of the mutant I96S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3PC0
 
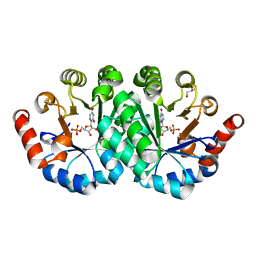 | | Crystal structure of the mutant V155S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
2HQG
 
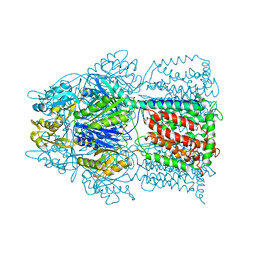 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
3PBV
 
 | | Crystal structure of the mutant I96T of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3TMU
 
 | | X-Ray Radiation Damage to HEWL Crystals soaked in 100mM Sodium Nitrate (Undosed) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Kmetko, J, Warkentin, M.A, Englich, U, Thorne, R.E. | | Deposit date: | 2011-08-31 | | Release date: | 2012-08-22 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Can radiation damage to protein crystals be reduced using small-molecule compounds?
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3TMW
 
 | | X-Ray Radiation Damage to HEWL Crystals soaked in 100mM Sodium Nitrate (Undosed) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Kmetko, J, Warkentin, M.A, Englich, U, Thorne, R.E. | | Deposit date: | 2011-08-31 | | Release date: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Can radiation damage to protein crystals be reduced using small-molecule compounds?
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3TMX
 
 | | X-Ray Radiation Damage to HEWL Crystals soaked in 100mM Sodium Nitrate (Dose=1.9MGy) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Kmetko, J, Warkentin, M.A, Englich, U, Thorne, R.E. | | Deposit date: | 2011-08-31 | | Release date: | 2012-08-22 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Can radiation damage to protein crystals be reduced using small-molecule compounds?
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2JVY
 
 | | Solution Structure of the EDA-ID-related C417F mutant of human NEMO zinc finger | | Descriptor: | NF-kappa-B essential modulator, ZINC ION | | Authors: | Cordier, F, Vinolo, E, Veron, M, Delepierre, M, Agou, F. | | Deposit date: | 2007-09-28 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of NEMO zinc finger and impact of an anhidrotic ectodermal dysplasia with immunodeficiency-related point mutation.
J.Mol.Biol., 377, 2008
|
|
