2E40
 
 | | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium in complex with gluconolactone | | Descriptor: | Beta-glucosidase, D-glucono-1,5-lactone | | Authors: | Nijikken, Y, Tsukada, T, Igarashi, K, Samejima, M, Fushinobu, S. | | Deposit date: | 2006-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium
Febs Lett., 581, 2007
|
|
4EUS
 
 | |
4CE5
 
 | | First crystal structure of an (R)-selective omega-transaminase from Aspergillus terreus | | Descriptor: | AT-OMEGATA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lyskowski, A, Gruber, C, Steinkellner, G, Schurmann, M, Schwab, H, Gruber, K, Steiner, K. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of an (R)-Selective Omega-Transaminase from Aspergillus Terreus
Plos One, 9, 2014
|
|
2CEZ
 
 | | Phosphorylation of the Cytoplasmic Tail of Tissue Factor and its Role in Modulating Structure and Binding Affinity | | Descriptor: | TISSUE FACTOR | | Authors: | Sen, M, Agrawal, S, Craft, J.W, Ruf, W, Legge, G.B. | | Deposit date: | 2006-02-13 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic Characterization of Successive Phosphorylation of the Tissue Factor Cytoplasmic Region.
Open Spectrosc J, 3, 2009
|
|
4CS4
 
 | | Catalytic domain of Pyrrolysyl-tRNA synthetase mutant Y306A, Y384F in complex with AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, 2-{[dihydroxy(4-aminoethylphenyl)-{4}-sulfanyl]amino}-3-hydroxypropanoic acid, MAGNESIUM ION, ... | | Authors: | Schmidt, M.J, Weber, A, Pott, M, Welte, W, Summerer, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells.
Chembiochem, 15, 2014
|
|
4CS2
 
 | | Catalytic domain of Pyrrolysyl-tRNA synthetase mutant Y306A, Y384F in its apo form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PYRROLYSINE--TRNA LIGASE | | Authors: | Schmidt, M.J, Weber, A, Pott, M, Welte, W, Summerer, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells.
Chembiochem, 15, 2014
|
|
2CEH
 
 | | Phosphorylation of the Cytoplasmic Tail of Tissue Factor and its Role in Modulating Structure and Binding Affinity | | Descriptor: | TISSUE FACTOR | | Authors: | Sen, M, Agrawal, S, Craft, J.W, Ruf, W, Legge, G.B. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic Characterization of Successive Phosphorylation of the Tissue Factor Cytoplasmic Region.
Open Spectrosc.J., 3, 2009
|
|
2CEF
 
 | | Phosphorylation of the Cytoplasmic Tail of Tissue Factor and its Role in Modulating Structure and Binding Affinity. | | Descriptor: | TISSUE FACTOR | | Authors: | Sen, M, Agrawal, S, Craft, J.W, Ruf, W, Legge, G.B. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-13 | | Last modified: | 2019-10-02 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic Characterization of Successive Phosphorylation of the Tissue Factor Cytoplasmic Region.
Open Spectrosc.J., 3, 2009
|
|
4DMC
 
 | |
2CFJ
 
 | | Phosphorylation of the Cytoplasmic Tail of Tissue Factor and its Role in Modulating Structure and Binding Affinity | | Descriptor: | TISSUE FACTOR | | Authors: | Sen, M, Agrawal, S, Craft, J.W, Ruf, W, Legge, G.B. | | Deposit date: | 2006-02-21 | | Release date: | 2007-03-20 | | Last modified: | 2019-10-02 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic Characterization of Successive Phosphorylation of the Tissue Factor Cytoplasmic Region.
Open Spectrosc.J., 3, 2009
|
|
4ER8
 
 | | Structure of the REP associates tyrosine transposase bound to a REP hairpin | | Descriptor: | DNA (32-MER), NICKEL (II) ION, TnpArep for protein | | Authors: | Messing, S.A.J, Ton-Hoang, B, Hickman, A.B, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The processing of repetitive extragenic palindromes: the structure of a repetitive extragenic palindrome bound to its associated nuclease.
Nucleic Acids Res., 40, 2012
|
|
2F99
 
 | | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity. | | Descriptor: | Aklanonic Acid methyl Ester Cyclase, AknH, SULFATE ION, ... | | Authors: | Kallio, P, Sultana, A, Neimi, J, Mantsala, P, Schneider, G. | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity.
J.Mol.Biol., 357, 2006
|
|
4DMF
 
 | |
4EX9
 
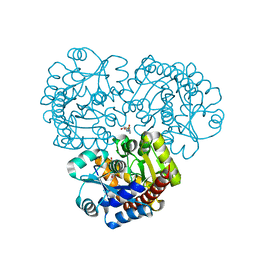 | | Crystal structure of the prealnumycin C-glycosynthase AlnA in complex with ribulose 5-phosphate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AlnA, CALCIUM ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2EWF
 
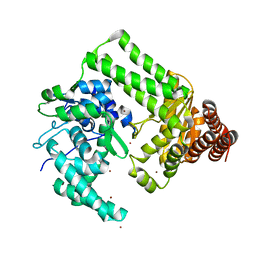 | | Crystal structure of the site-specific DNA nickase N.BspD6I | | Descriptor: | BROMIDE ION, Nicking endonuclease N.BspD6I | | Authors: | Kachalova, G.S, Bartunik, H.D, Artyukh, R.I, Rogulin, E.A, Perevyazova, T.A, Zheleznaya, L.A, Matvienko, N.I. | | Deposit date: | 2005-11-03 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit.
J.Mol.Biol., 384, 2008
|
|
4DLN
 
 | |
4DM7
 
 | |
4D1Q
 
 | | Hermes transposase bound to its terminal inverted repeat | | Descriptor: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | Authors: | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | Deposit date: | 2014-05-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
2FHR
 
 | | Trypanosoma Rangeli Sialidase In Complex With 2,3- Difluorosialic Acid (Covalent Intermediate) | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Amaya, M.F, Alzari, P.M, Buschiazzo, A. | | Deposit date: | 2005-12-26 | | Release date: | 2006-01-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Kinetic Analysis of Two Covalent Sialosyl-Enzyme Intermediates on Trypanosoma rangeli Sialidase.
J.Biol.Chem., 281, 2006
|
|
4EX7
 
 | | Crystal structure of the alnumycin P phosphatase in complex with free phosphate | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EX8
 
 | | Crystal structure of the prealnumycin C-glycosynthase AlnA | | Descriptor: | AlnA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4C74
 
 | | Phenylacetone monooxygenase: Reduced enzyme in complex with APADP | | Descriptor: | 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, HEXAETHYLENE GLYCOL, ... | | Authors: | Martinoli, C, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2013-09-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Beyond the Protein Matrix: Probing Cofactor Variants in a Baeyer-Villiger Oxygenation Reaction.
Acs Catal., 3, 2013
|
|
4CCY
 
 | | Crystal structure of carboxylesterase CesB (YbfK) from Bacillus subtilis | | Descriptor: | CARBOXYLESTERASE YBFK, SODIUM ION | | Authors: | Rozeboom, H.J, Godinho, L.F, Nardini, M, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structures of Two Bacillus Carboxylesterases with Different Enantioselectivities.
Biochim.Biophys.Acta, 1844, 2014
|
|
2F40
 
 | |
2G7E
 
 | | The 1.6 A crystal structure of Vibrio cholerae extracellular endonuclease I | | Descriptor: | CHLORIDE ION, Endonuclease I | | Authors: | Altermark, B, Smalaas, A.O, Willassen, N.P, Helland, R. | | Deposit date: | 2006-02-28 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of Vibrio cholerae extracellular endonuclease I reveals the presence of a buried chloride ion.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
