5Y88
 
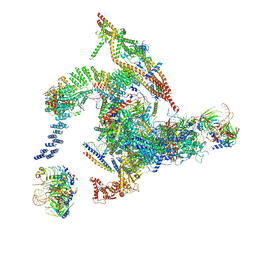 | | Cryo-EM structure of the intron-lariat spliceosome ready for disassembly from S.cerevisiae at 3.5 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat, ... | | Authors: | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | Deposit date: | 2017-08-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae
Cell(Cambridge,Mass.), 171, 2017
|
|
5XJC
 
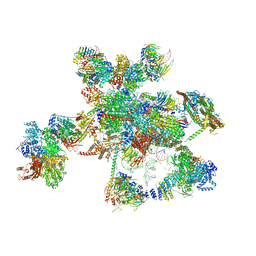 | | Cryo-EM structure of the human spliceosome just prior to exon ligation at 3.6 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Yan, C, Hang, J, Finci, I.L, Lei, J, Shi, Y. | | Deposit date: | 2017-04-30 | | Release date: | 2017-07-05 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An Atomic Structure of the Human Spliceosome
Cell, 169, 2017
|
|
5X8R
 
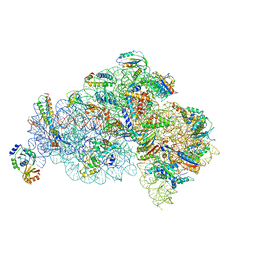 | | Structure of the 30S small subunit of chloroplast ribosome from spinach | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, chloroplastic, ... | | Authors: | Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation
Nucleic Acids Res., 45, 2017
|
|
5X8P
 
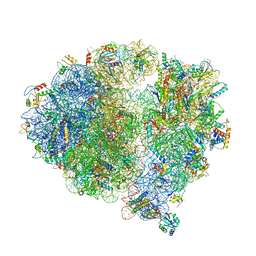 | | Structure of the 70S chloroplast ribosome from spinach | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation
Nucleic Acids Res., 45, 2017
|
|
5X3Z
 
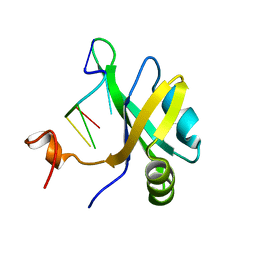 | | Solution structure of musashi1 RBD2 in complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | Descriptor: | RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5WWG
 
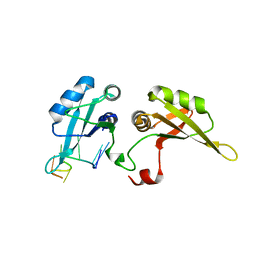 | | Crystal structure of hnRNPA2B1 in complex with AAGGACUUGC | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*AP*GP*GP*AP*CP*UP*U)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
5WWF
 
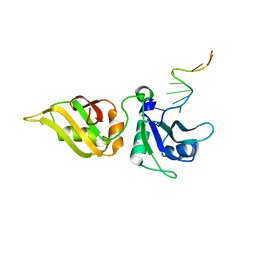 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
5WWE
 
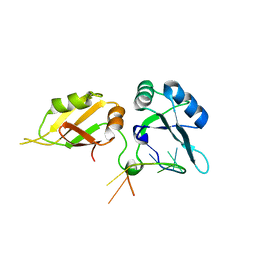 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*GP*GP*GP*AP*CP*UP*AP*GP*A)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
5WU6
 
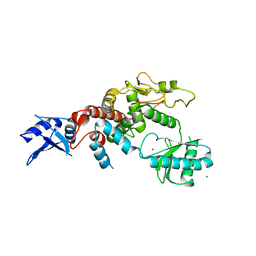 | | Crystal structure of apo human Tut1, form IV | | Descriptor: | MAGNESIUM ION, Speckle targeted PIP5K1A-regulated poly(A) polymerase | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2016-12-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Crystal structures of U6 snRNA-specific terminal uridylyltransferase
Nat Commun, 8, 2017
|
|
5WSG
 
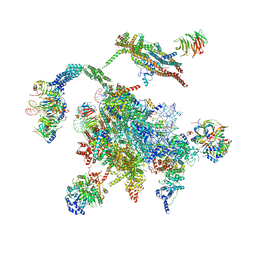 | | Cryo-EM structure of the Catalytic Step II spliceosome (C* complex) at 4.0 angstrom resolution | | Descriptor: | 3'-exon-intron, 3'-intron-lariat, 5'-exon, ... | | Authors: | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of a yeast step II catalytically activated spliceosome
Science, 355, 2017
|
|
5WPA
 
 | | Structure of human SFPQ/PSPC1 heterodimer | | Descriptor: | Paraspeckle component 1, Splicing factor, proline- and glutamine-rich | | Authors: | Lee, M. | | Deposit date: | 2017-08-04 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of a SFPQ/PSPC1 heterodimer provides insights into preferential heterodimerization of human DBHS family proteins.
J. Biol. Chem., 293, 2018
|
|
5W0H
 
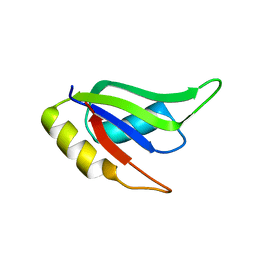 | |
5W0G
 
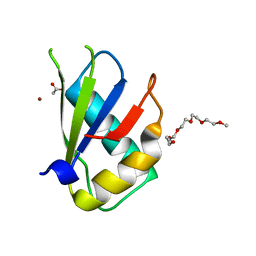 | | Structure of U2AF65 (U2AF2) RRM1 at 1.07 resolution | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, Splicing factor U2AF 65 kDa subunit, ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2017-05-30 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Cancer-Associated Mutations Mapped on High-Resolution Structures of the U2AF2 RNA Recognition Motifs.
Biochemistry, 56, 2017
|
|
5VSU
 
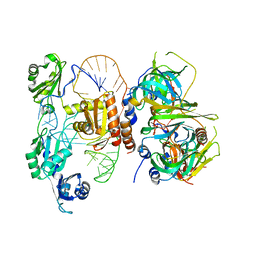 | | Structure of yeast U6 snRNP with 2'-phosphate terminated U6 RNA | | Descriptor: | Saccharomyces cerevisiae strain T8 chromosome XII sequence, U4/U6 snRNA-associated-splicing factor PRP24, U6 snRNA-associated Sm-like protein LSm2, ... | | Authors: | Montemayor, E.J. | | Deposit date: | 2017-05-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Architecture of the U6 snRNP reveals specific recognition of 3'-end processed U6 snRNA.
Nat Commun, 9, 2018
|
|
5UZM
 
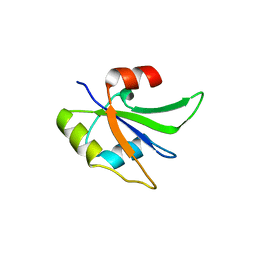 | | Crystal structure of Glorund qRRM2 domain | | Descriptor: | AT27789p | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
5UZG
 
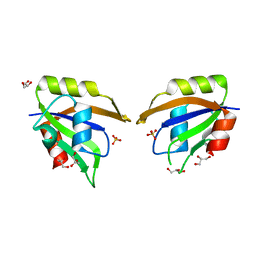 | | Crystal structure of Glorund qRRM1 domain | | Descriptor: | AT27789p, GLYCEROL, SULFATE ION | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
5TKZ
 
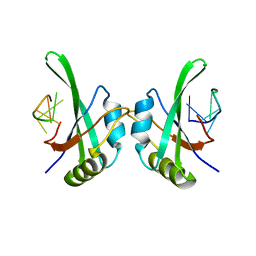 | | MEC-8 N-terminal RRM bound to tandem GCAC ligand | | Descriptor: | DNA (5'-D(*AP*GP*CP*AP*CP*AP*TP*TP*TP*TP*TP*TP*TP*TP*AP*GP*CP*AP*CP*A)-3'), Mec-8 protein | | Authors: | Soufari, H, Mackereth, C.D. | | Deposit date: | 2016-10-10 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Conserved binding of GCAC motifs by MEC-8, couch potato, and the RBPMS protein family.
RNA, 23, 2017
|
|
5TF6
 
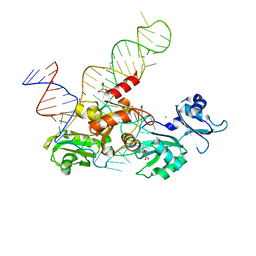 | | Structure and conformational plasticity of the U6 small nuclear ribonucleoprotein core | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Montemayor, E.J, Brow, D.A, Butcher, S.E. | | Deposit date: | 2016-09-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and conformational plasticity of the U6 small nuclear ribonucleoprotein core.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5TBX
 
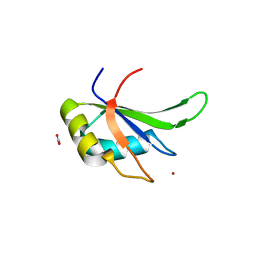 | | hnRNP A18 RNA Recognition Motif | | Descriptor: | ACETATE ION, Cold-inducible RNA-binding protein, NICKEL (II) ION | | Authors: | Coburn, K.M, Melville, Z, Aligholizadeh, E, Roth, B.M, Varney, K.M, Weber, D.J. | | Deposit date: | 2016-09-13 | | Release date: | 2017-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Crystal structure of the human heterogeneous ribonucleoprotein A18 RNA-recognition motif.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5T9P
 
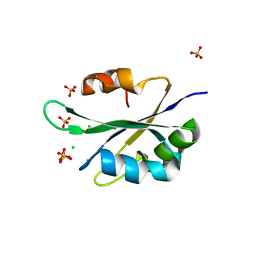 | |
5SZW
 
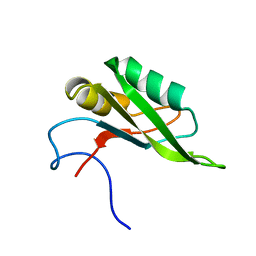 | | NMR solution structure of the RRM1 domain of the post-transcriptional regulator HuR | | Descriptor: | ELAV-like protein 1 | | Authors: | Lixa, C, Mujo, A, Jendiroba, K.A, Almeida, F.C.L, Lima, L.M.T.R, Pinheiro, A.S. | | Deposit date: | 2016-08-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oligomeric transition and dynamics of RNA binding by the HuR RRM1 domain in solution.
J. Biomol. NMR, 72, 2018
|
|
5OSG
 
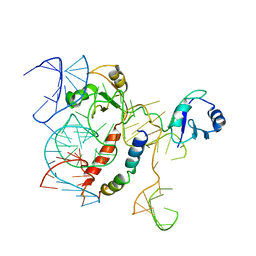 | | Structure of KSRP in context of Leishmania donovani 80S | | Descriptor: | 18S rRNA, 40S ribosomal protein S6, RNA binding protein, ... | | Authors: | Brito Querido, J, Mancera-Martinez, E, Vicens, Q, Bochler, A, Chicher, J, Simonetti, A, Hashem, Y. | | Deposit date: | 2017-08-17 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein.
Structure, 25, 2017
|
|
5OPT
 
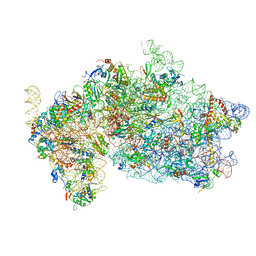 | | Structure of KSRP in context of Trypanosoma cruzi 40S | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, putative, ... | | Authors: | Brito Querido, J, Mancera-Martinez, E, Vicens, Q, Bochler, A, Chicher, J, Simonetti, A, Hashem, Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein.
Structure, 25, 2017
|
|
5OOB
 
 | |
