8AT9
 
 | | Small molecule stabilizer for ERalpha and 14-3-3 (1080269) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-(1-phenylazanylcyclobutyl)carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-22 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AU2
 
 | | Small molecular stabilizer for ERalpha and 14-3-3 (1080293) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[2-[1-[4-[(4-chlorophenyl)amino]piperidin-4-yl]carbonylpiperidin-4-yl]ethyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-25 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AV3
 
 | | Small molecular stabilizer for ERalpha and 14-3-3 (1075299) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[1-(4-chloranylphenoxy)-4,4-bis(fluoranyl)cyclohexyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-26 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AXE
 
 | | Small molecule stabilizer for ERalpha and 14-3-3 (1074210) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[2-[1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]piperidin-4-yl]ethyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-08-31 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ALV
 
 | | Small molecular stabilizer for ERalpha and 14-3-3 (1076403) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-[1-[(4-chlorophenyl)amino]cyclohexyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ARX
 
 | | Small molecular stabilizer for ERalpha and 14-3-3sigma (1074378) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-[1-(4-chloranylphenoxy)cyclopropyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AI0
 
 | | Small molecular stabilizer for ERalpha and 14-3-3sigma (1080268) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-(2-methyl-2-phenylazanyl-propanoyl)piperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8WRJ
 
 | | glycosyltransferase UGT74AN3 | | Descriptor: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Huang, W, Long, F. | | Deposit date: | 2023-10-15 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8BBS
 
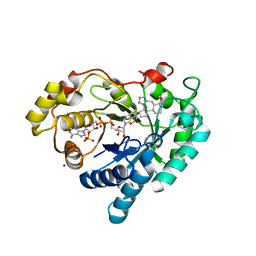 | | Structure of AKR1C3 in complex with a bile acid fused tetrazole inhibitor | | Descriptor: | (4~{R})-4-[(1~{R},2~{S},5~{R},6~{R},13~{S},14~{S},17~{R},19~{R})-6,14-dimethyl-17-oxidanyl-7,8,9,10-tetrazapentacyclo[11.8.0.0^{2,6}.0^{7,11}.0^{14,19}]henicosa-8,10-dien-5-yl]pentanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Petri, E.T, Skerlova, J, Marinovic, M, Brynda, J, Kugler, M, Skoric, D, Bekic, S, Celic, A.S, Rezacova, P. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray structure of human aldo-keto reductase 1C3 in complex with a bile acid fused tetrazole inhibitor: experimental validation, molecular docking and structural analysis.
Rsc Med Chem, 14, 2023
|
|
7O3F
 
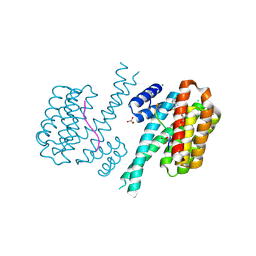 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-117 | | Descriptor: | 1-methyl-4-(4-methylphenyl)sulfonyl-1,4-diazepane, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O6J
 
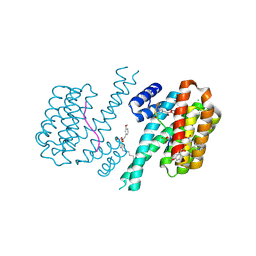 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-083 | | Descriptor: | (5-methanoyl-2-nitro-phenyl) 2-(3-methoxyphenyl)ethanoate, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O3A
 
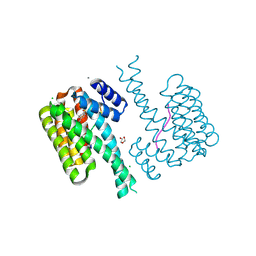 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-046 | | Descriptor: | 1-[4-(hydroxymethyl)phenyl]sulfonylpiperidin-4-ol, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O6M
 
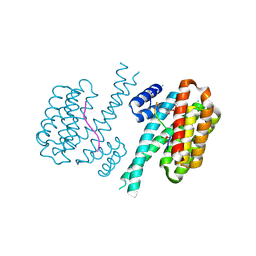 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-097 | | Descriptor: | (5-methanoyl-2-nitro-phenyl) 1-methylpyrazole-4-sulfonate, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O6K
 
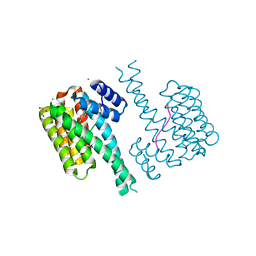 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-080 | | Descriptor: | (5-methanoyl-2-nitro-phenyl) 3-chloranylbenzoate, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
8BZF
 
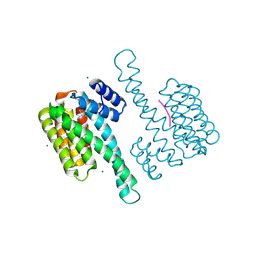 | | FC-J acetonide stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, Fusicoccin J-acetonide, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
7O6O
 
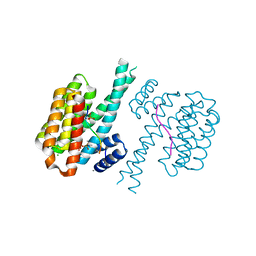 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-096 | | Descriptor: | (5-methanoyl-2-nitro-phenyl) propane-2-sulfonate, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
8B39
 
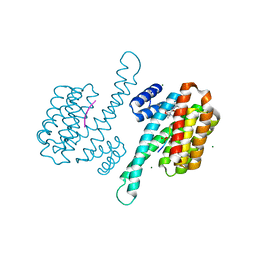 | | Small molecular stabilizer for ERalpha and 14-3-3 (1080299) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[(2~{S},6~{R})-4-[(4-chlorophenyl)amino]-2,6-dimethyl-oxan-4-yl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8BZD
 
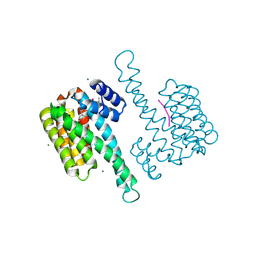 | | deAc-FC stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, De-acetylated Fusicoccin, ERalpha peptide, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
8AH2
 
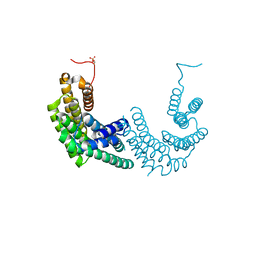 | | Crystal structure of human 14-3-3 zeta fused to the NPM1 peptide including phosphoserine-48 | | Descriptor: | 14-3-3 protein zeta/delta,Nucleophosmin | | Authors: | Boyko, K.M, Kapitonova, A.A, Tugaeva, K.V, Varfolomeeva, L.A, Sluchanko, N.N. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the recognition by 14-3-3 proteins of a conditional binding site within the oligomerization domain of human nucleophosmin.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
6TLF
 
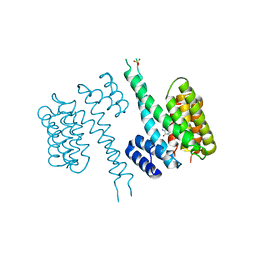 | | human 14-3-3 sigma isoform in complex with IMP | | Descriptor: | 14-3-3 protein sigma, INOSINIC ACID, SULFATE ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-12-02 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of Phosphate-Containing Compounds as New Inhibitors of 14-3-3/c-Abl Protein-Protein Interaction.
Acs Chem.Biol., 15, 2020
|
|
8A9G
 
 | | Binary complex of 14-3-3 zeta Glucocorticoid Receptor (GR) pT524 peptide stabilised by (R)-para chloropyrrolidone1 | | Descriptor: | 14-3-3 protein zeta/delta, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(2~{R})-3-(4-chlorophenyl)carbonyl-2-(4-nitrophenyl)-4-oxidanyl-5-oxidanylidene-2~{H}-pyrrol-1-yl]-2-oxidanyl-benzoic acid, ... | | Authors: | Munier, C.C, Edman, K, Perry, M.W.D, Ottmann, C. | | Deposit date: | 2022-06-28 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Designing Selective Drug-like Molecular Glues for the Glucocorticoid Receptor/14-3-3 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
8BWH
 
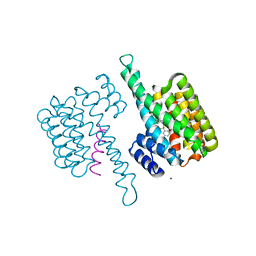 | |
8BWE
 
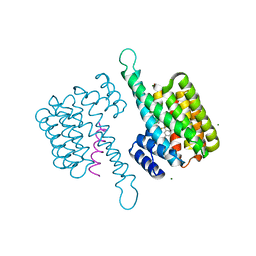 | |
8C1Y
 
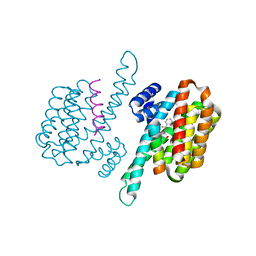 | | Small molecule stabilizer for 14-3-3/ChREBP (Cmd 30) | | Descriptor: | 14-3-3 protein sigma, Carbohydrate-responsive element-binding protein, [2-[2-[[2,2-bis(fluoranyl)-2-phenyl-ethyl]amino]-2-oxidanylidene-ethoxy]phenyl]phosphonic acid | | Authors: | Pennings, M.A.M, Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular glues of the regulatory ChREBP/14-3-3 complex protect beta cells from glucolipotoxicity.
Biorxiv, 2024
|
|
7Z2Z
 
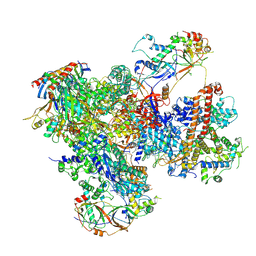 | | Structure of yeast RNA Polymerase III-DNA-Ty1 integrase complex (Pol III-DNA-IN1) at 3.1 A | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Nguyen, P.Q, Fernandez-Tornero, C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
