2GKT
 
 | |
2VR2
 
 | | Human Dihydropyrimidinase | | Descriptor: | CHLORIDE ION, DIHYDROPYRIMIDINASE, ZINC ION | | Authors: | Welin, M, Karlberg, T, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Human Dihydropyrimidinase
To be Published
|
|
4B6W
 
 | | Architecture of Trypanosoma brucei Tubulin-Binding cofactor B | | Descriptor: | 1,2-ETHANEDIOL, TUBULIN-SPECIFIC CHAPERONE | | Authors: | Fleming, J.R, Morgan, R.E, Fyfe, P.K, Kelly, S.M, Hunter, W.N. | | Deposit date: | 2012-08-15 | | Release date: | 2012-08-22 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Architecture of Trypanosoma Brucei Tubulin-Binding Cofactor B and Implications for Function.
FEBS J., 280, 2013
|
|
2GKR
 
 | | Crystal structure of the N-terminally truncated OMTKY3-del(1-5) | | Descriptor: | CHLORIDE ION, Ovomucoid | | Authors: | Lee, T.W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-04-03 | | Release date: | 2007-02-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural Insights into the Non-additivity Effects in the Sequence-to-Reactivity Algorithm for Serine Peptidases and their Inhibitors.
J.Mol.Biol., 367, 2007
|
|
2R9Y
 
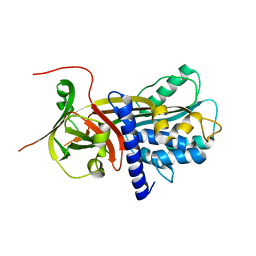 | | Structure of antiplasmin | | Descriptor: | Alpha-2-antiplasmin | | Authors: | Law, R.H.P, Sofian, T, Kan, W.T, Horvath, A.J, Hitchen, C.R, Langendorf, C.G, Buckle, A.M, Whisstock, J.C, Coughlin, P.B. | | Deposit date: | 2007-09-14 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray crystal structure of the fibrinolysis inhibitor {alpha}2-antiplasmin
Blood, 111, 2008
|
|
3ZVA
 
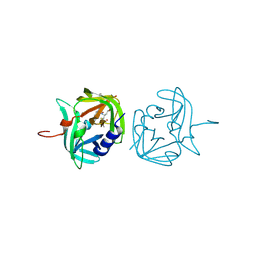 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 75 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3TEX
 
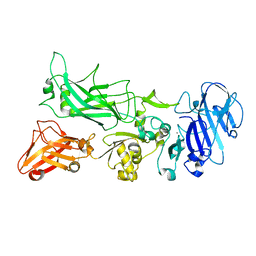 | |
3TEW
 
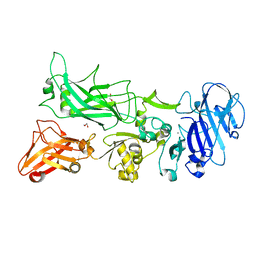 | |
2WPO
 
 | | HCMV protease inhibitor complex | | Descriptor: | (2S)-2-(3,3-dimethylbutanoylamino)-N-[(2S)-1-[[(2S,3S)-3-hydroxy-4-[(4-iodophenyl)methylamino]-4-oxo-butan-2-yl]amino]- 1,4-dioxo-4-pyrrol-1-yl-butan-2-yl]-3,3-dimethyl-butanamide, HUMAN CYTOMEGALOVIRUS PROTEASE | | Authors: | Tong, L, Qian, C, Massariol, M.-J, Deziel, R, Yoakim, C, Lagace, L. | | Deposit date: | 1998-08-04 | | Release date: | 1999-08-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conserved mode of peptidomimetic inhibition and substrate recognition of human cytomegalovirus protease.
Nat.Struct.Biol., 5, 1998
|
|
1WQS
 
 | | Crystal structure of Norovirus 3C-like protease | | Descriptor: | 3C-like protease, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Nakamura, K, Someya, Y, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-10-01 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A norovirus protease structure provides insights into active and substrate binding site integrity
J.Virol., 79, 2005
|
|
3UFA
 
 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | Descriptor: | CHLORIDE ION, N-(3-carboxypropanoyl)-L-valyl-N-[(1S)-2-phenyl-1-phosphonoethyl]-L-prolinamide, Serine protease splA | | Authors: | Zdzalik, M, Pietrusewicz, E, Pustelny, K, Stec-Niemczyk, J, Popowicz, G.M, Potempa, J, Oleksyszyn, J, Dubin, G. | | Deposit date: | 2011-10-31 | | Release date: | 2013-01-23 | | Last modified: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
3SY3
 
 | | GBAA_1210 protein, a putative adenylate cyclase, from Bacillus anthracis | | Descriptor: | GBAA_1210 protein, PHOSPHATE ION | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-15 | | Release date: | 2011-07-27 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | GBAA_1210 protein, a putative adenylate cyclase, from Bacillus anthracis.
To be Published
|
|
3QXX
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GDP and 8-aminocaprylic acid | | Descriptor: | 1,2-ETHANEDIOL, 8-aminooctanoic acid, Dethiobiotin synthetase, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXH
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ADP and 8-aminocaprylic acid | | Descriptor: | 1,2-ETHANEDIOL, 8-aminooctanoic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Minor, C, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXS
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ANP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, MAGNESIUM ION, ... | | Authors: | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Jablonska, K, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXC
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXJ
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GTP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QY0
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3Q8A
 
 | | Crystal structure of WT Protective Antigen (pH 5.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Rajapaksha, M, Lovell, S, Janowiak, B.E, Andra, K.K, Battaile, K.P, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.129 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
1UOQ
 
 | |
3Q8B
 
 | | Crystal structure of WT Protective Antigen (pH 9.0) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
1UOO
 
 | |
3V3M
 
 | | Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease in Complex with N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide inhibitor. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | | Authors: | Jacobs, J, Grum-Tokars, V, Zhou, Y, Turlington, M, Saldanha, S.A, Chase, P, Eggler, A, Dawson, E.S, Baez-Santos, Y.M, Tomar, S, Mielech, A.M, Baker, S.C, Lindsley, C.W, Hodder, P, Mesecar, A, Stauffer, S.R. | | Deposit date: | 2011-12-13 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery, Synthesis, And Structure-Based Optimization of a Series of N-(tert-Butyl)-2-(N-arylamido)-2-(pyridin-3-yl) Acetamides (ML188) as Potent Noncovalent Small Molecule Inhibitors of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease.
J.Med.Chem., 56, 2013
|
|
3TEZ
 
 | |
3TEY
 
 | |
