7LKS
 
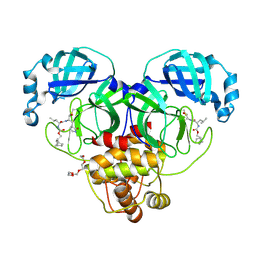 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2f | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKT
 
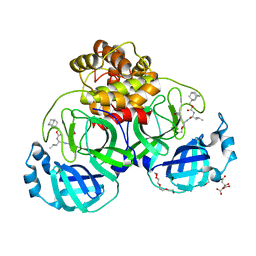 | | 1.50 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2k | | Descriptor: | (1R,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKW
 
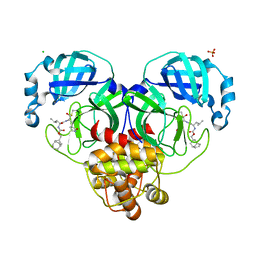 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3d (deuterated analog of inhibitor 3c) | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
2XAK
 
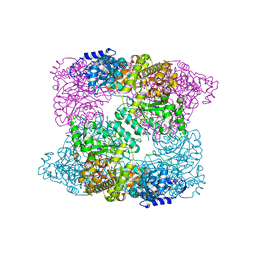 | | Ribonucleotide reductase Y730NO2Y modified R1 subunit of E. coli | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Yokoyama, K, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Site-Specific Incorporation of 3-Nitrotyrosine as a Probe of Pk(A) Perturbation of Redox-Active Tyrosines in Ribonucleotide Reductase.
J.Am.Chem.Soc., 132, 2010
|
|
6CEL
 
 | | CBH1 (E212Q) CELLOPENTAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
1ZJY
 
 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NADH | | Descriptor: | (1R)-1-PHENYLETHANOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
5OWA
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ054 | | Descriptor: | Peregrin, ~{N}-[4-[[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]methyl]-1,3-thiazol-2-yl]-2,4-dimethyl-1,3-oxazole-5-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-08-31 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
5XWD
 
 | | Crystal structure of the complex of 059-152-Fv and EGFR-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Matsuda, T, Ito, T, Shirouzu, M. | | Deposit date: | 2017-06-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Cell-free synthesis of functional antibody fragments to provide a structural basis for antibody-antigen interaction
PLoS ONE, 13, 2018
|
|
2FR8
 
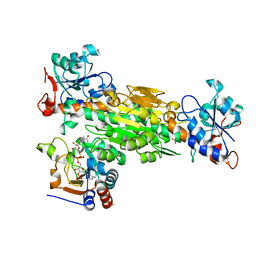 | | Structure of transhydrogenase (dI.R127A.NAD+)2(dIII.NADP+)1 asymmetric complex | | Descriptor: | NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Brondijk, T.H, van Boxel, G.I, Mather, O.C, Quirk, P.G, White, S.A, Jackson, J.B. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Role of Invariant Amino Acid Residues at the Hydride Transfer Site of Proton-translocating Transhydrogenase.
J.Biol.Chem., 281, 2006
|
|
1WCF
 
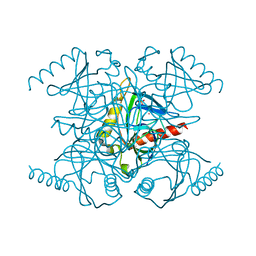 | |
3SDV
 
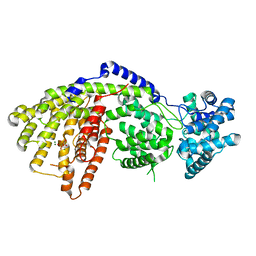 | | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production | | Descriptor: | (1-hydroxyethane-1,1-diyl)bis(phosphonic acid), Alpha-bisabolene synthase, CHLORIDE ION, ... | | Authors: | McAndrew, R.P, Peralta-Yahya, P.P, DeGiovanni, A, Pereira, J.H, Hadi, M.Z, Keasling, J.D, Adams, P.D. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production.
Structure, 19, 2011
|
|
3S8B
 
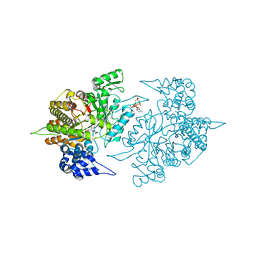 | | Structure of Yeast Ribonucleotide Reductase 1 with AMPPNP and CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical basis of lethal mutant R293A of yeast ribonucleotide reductase
To be Published
|
|
7F9O
 
 | | PSI-NDH supercomplex of Barley | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2021-07-04 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
2WP9
 
 | | Crystal structure of the E. coli succinate:quinone oxidoreductase (SQR) SdhB His207Thr mutant | | Descriptor: | 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-08-03 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Perturbation of the quinone-binding site of complex II alters the electronic properties of the proximal [3Fe-4S] iron-sulfur cluster.
J. Biol. Chem., 286, 2011
|
|
7K1E
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_88646 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 3,4-difluoro-N-[(1S)-1-{1-[(2R)-4-(hydroxyamino)-4-oxo-1-(5,6,7,8-tetrahydronaphthalen-2-yl)butan-2-yl]-1H-1,2,3-triazol-4-yl}ethyl]benzamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 3
To Be Published
|
|
3LZ6
 
 | | Guinea Pig 11beta hydroxysteroid dehydrogenase with PF-877423 | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-adamantan-2-yl-1-ethyl-D-prolinamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pauly, T.A. | | Deposit date: | 2010-03-01 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The development and SAR of pyrrolidine carboxamide 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7K9O
 
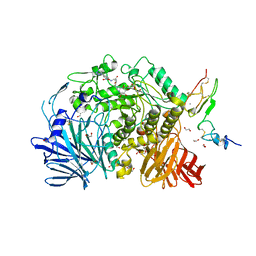 | | Co-crystal structure of alpha glucosidase with compound 3 | | Descriptor: | (1~{S},2~{S},3~{R},4~{S},5~{S})-1-(hydroxymethyl)-5-[6-[[2-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]-4-(1,2,3,4-tetrazol-1-yl)phenyl]amino]hexylamino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
2CEP
 
 | |
5ZCP
 
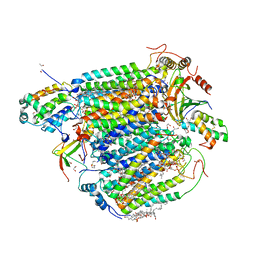 | | azide-bound cytochrome c oxidase structure determined using the crystals exposed to 20 mM azide solution for 2 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-02-19 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
1RC2
 
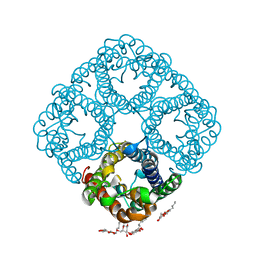 | | 2.5 Angstrom Resolution X-ray Structure of Aquaporin Z | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, Aquaporin Z | | Authors: | Savage, D.F, Egea, P.F, Robles, Y.C, O'Connell III, J.D, Stroud, R.M. | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture and selectivity in aquaporins: 2.5 a X-ray structure of aquaporin Z
Plos Biol., 1, 2003
|
|
2I5P
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase isoform 1 from K. marxianus | | Descriptor: | BETA-MERCAPTOETHANOL, Glyceraldehyde-3-phosphate dehydrogenase 1, alpha-D-glucopyranose | | Authors: | Ferreira-da-Silva, F, Pereira, P.J.B, Gales, L, Moradas-Ferreira, P, Damas, A.M. | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal and Solution Structures of Glyceraldehyde-3-phosphate Dehydrogenase Reveal Different Quaternary Structures.
J.Biol.Chem., 281, 2006
|
|
3SAI
 
 | |
3S74
 
 | |
5Z86
 
 | | azide-bound cytochrome c oxidase structure determined using the crystals exposed to 20 mM azide solution for 3 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-01-31 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
1GGE
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, NATIVE STRUCTURE AT 1.9 A RESOLUTION. | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, PROTEIN (CATALASE HPII) | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-16 | | Release date: | 2000-08-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
