4V4G
 
 | | Crystal structure of five 70s ribosomes from Escherichia Coli in complex with protein Y. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | Authors: | Vila-Sanjurjo, A, Schuwirth, B.S, Hau, C.W, Cate, J.H. | | Deposit date: | 2004-10-06 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (11.5 Å) | | Cite: | Structural basis for the control of translation initiation during stress.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4V99
 
 | |
6EKC
 
 | | Crystal structure of the BSD2 homolog of Arabidopsis thaliana bound to the octameric assembly of RbcL from Thermosynechococcus elongatus | | Descriptor: | DnaJ/Hsp40 cysteine-rich domain superfamily protein, Ribulose bisphosphate carboxylase large chain, ZINC ION | | Authors: | Aigner, H, Wilson, R.H, Bracher, A, Calisse, L, Bhat, J.Y, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-09-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2.
Science, 358, 2017
|
|
3MYI
 
 | |
3QHF
 
 | |
3QHZ
 
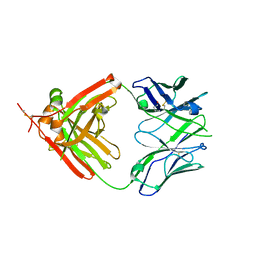 | | Crystal Structure of human anti-influenza Fab 2D1 | | Descriptor: | Human monoclonal antibody del2D1, Fab Heavy Chain, Fab Light Chain | | Authors: | Ekiert, D.C, Wilson, I.A. | | Deposit date: | 2011-01-26 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An insertion mutation that distorts antibody binding site architecture enhances function of a human antibody.
MBio, 2, 2011
|
|
5IBU
 
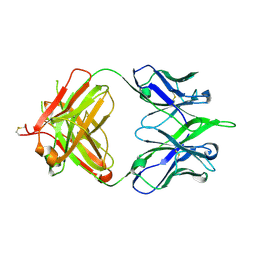 | | 6652 Fab (unbound) | | Descriptor: | 6652 Heavy Chain, 6652 Light Chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2016-02-22 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain.
Nat. Med., 22, 2016
|
|
5JS9
 
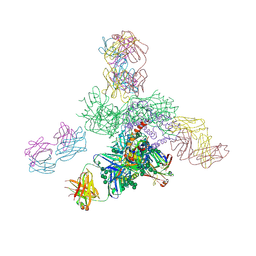 | | Uncleaved prefusion optimized gp140 trimer with an engineered 8-residue HR1 turn bound to broadly neutralizing antibodies 8ANC195 and PGT128 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2016-05-07 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (6.918 Å) | | Cite: | Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability.
Nat Commun, 7, 2016
|
|
6H9Z
 
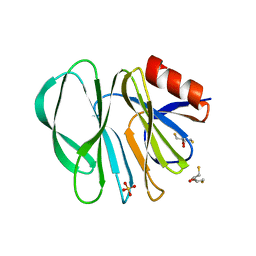 | | Molecular bases of histo-blood group antigen recognition by the most common human rotavirus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Outer capsid protein VP4, SULFATE ION | | Authors: | Ciges-Tomas, J.R, Gozalbo-Rovira, R, Vila-Vicent, S, Buesa, J, Santiso-Bellon, C, Monedero, V, Yebra, M.J, Rodriguez-Diaz, J, Marina, A. | | Deposit date: | 2018-08-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens.
Plos Pathog., 15, 2019
|
|
5JSA
 
 | | Uncleaved prefusion optimized gp140 trimer with an engineered 10-residue HR1 turn bound to broadly neutralizing antibodies 8ANC195 and PGT128 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2016-05-07 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (6.308 Å) | | Cite: | Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability.
Nat Commun, 7, 2016
|
|
7NVG
 
 | | Salmonella flagellar basal body refined in C1 map | | Descriptor: | Basal-body rod modification protein FlgD, Flagellar L-ring protein, Flagellar M-ring protein, ... | | Authors: | Johnson, S, Furlong, E, Lea, S.M. | | Deposit date: | 2021-03-15 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
6H9Y
 
 | | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Outer capsid protein VP4, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ciges-Tomas, J.R, Gozalbo-Rovira, R, Vila-Vicent, S, Buesa, J, Santiso-Bellon, C, Monedero, V, Yebra, M.J, Rodriguez-Diaz, J, Marina, A. | | Deposit date: | 2018-08-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens.
Plos Pathog., 15, 2019
|
|
6H9W
 
 | | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens | | Descriptor: | CITRIC ACID, ISOPROPYL ALCOHOL, Outer capsid protein VP4, ... | | Authors: | Ciges-Tomas, J.R, Gozalbo-Rovira, R, Vila-Vicent, S, Buesa, J, Santiso-Bellon, C, Monedero, V, Yebra, M.J, Rodriguez-Diaz, J, Marina, A. | | Deposit date: | 2018-08-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens.
Plos Pathog., 15, 2019
|
|
6HA0
 
 | | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens | | Descriptor: | Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ciges-Tomas, J.R, Gozalbo-Rovira, R, Vila-Vicent, S, Buesa, J, Santiso-Bellon, C, Monedero, V, Yebra, M.J, Rodriguez-Diaz, J, Marina, A. | | Deposit date: | 2018-08-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens.
Plos Pathog., 15, 2019
|
|
7K63
 
 | |
5C7K
 
 | | Crystal structure BG505 SOSIP gp140 HIV-1 Env trimer bound to broadly neutralizing antibodies PGT128 and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 8ANC195 heavy chain, ... | | Authors: | Kong, L, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.6019 Å) | | Cite: | Complete epitopes for vaccine design derived from a crystal structure of the broadly neutralizing antibodies PGT128 and 8ANC195 in complex with an HIV-1 Env trimer.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5FYK
 
 | | Crystal Structure at 3.7 A Resolution of Fully Glycosylated HIV-1 Clade B JR-FL SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122, 35O22 and VRC01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.107 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
5FYL
 
 | | Crystal Structure at 3.7 A Resolution of Fully Glycosylated HIV-1 Clade A BG505 SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 ANTIBODY FAB HEAVY CHAIN, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
6WLA
 
 | | Antigen binding fragment of ch128.1 | | Descriptor: | Fab ch128.1 heavy chain, Fab ch128.1 light chain, GLYCEROL | | Authors: | Helguera, G, Rodriguez, J.A, Sawaya, M, Cascio, D, Zink, S, Ziegenbein, J, Short, C. | | Deposit date: | 2020-04-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Host receptor-targeted therapeutic approach to counter pathogenic New World mammarenavirus infections.
Nat Commun, 13, 2022
|
|
2IJH
 
 | | Crystal structure analysis of ColE1 ROM mutant F14W | | Descriptor: | Regulatory protein rop | | Authors: | Ladner, J.E. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
5T3X
 
 | |
2IJJ
 
 | | Crystal structure analysis of ColE1 ROM mutant F14Y | | Descriptor: | Regulatory protein rop | | Authors: | Ladner, J.E. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
2IJK
 
 | | Structure of a Rom protein dimer at 1.55 angstrom resolution | | Descriptor: | Regulatory protein rop | | Authors: | Struble, E.B. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
2IJI
 
 | | Structure of F14H mutant of ColE1 Rom protein | | Descriptor: | Regulatory protein rop | | Authors: | Struble, E.B. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
4PY5
 
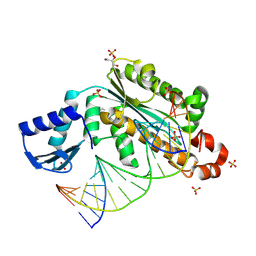 | | Thermovibrio ammonificans RNase H3 in complex with 19-mer RNA/DNA | | Descriptor: | 5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*GP*CP*AP*CP*TP*C)-3', 5'-R(*GP*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3', GLYCEROL, ... | | Authors: | Figiel, M, Nowotny, M. | | Deposit date: | 2014-03-26 | | Release date: | 2014-07-30 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of RNase H3-substrate complex reveals parallel evolution of RNA/DNA hybrid recognition.
Nucleic Acids Res., 42, 2014
|
|
